|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Denisenko TV, Budkevich IN and Zhivotovsky
B: Cell death-based treatment of lung adenocarcinoma. Cell Death
Dis. 9(117)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kim HC, Jung CY, Cho DG, Jeon JH, Lee JE,
Ahn JS, Kim SJ, Kim Y, Kim YC, Kim JE, et al: Clinical
characteristics and prognostic factors of lung cancer in Korea: A
pilot study of data from the Korean nationwide lung cancer
registry. Tuberc Respir Dis (Seoul). 82:118–125. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
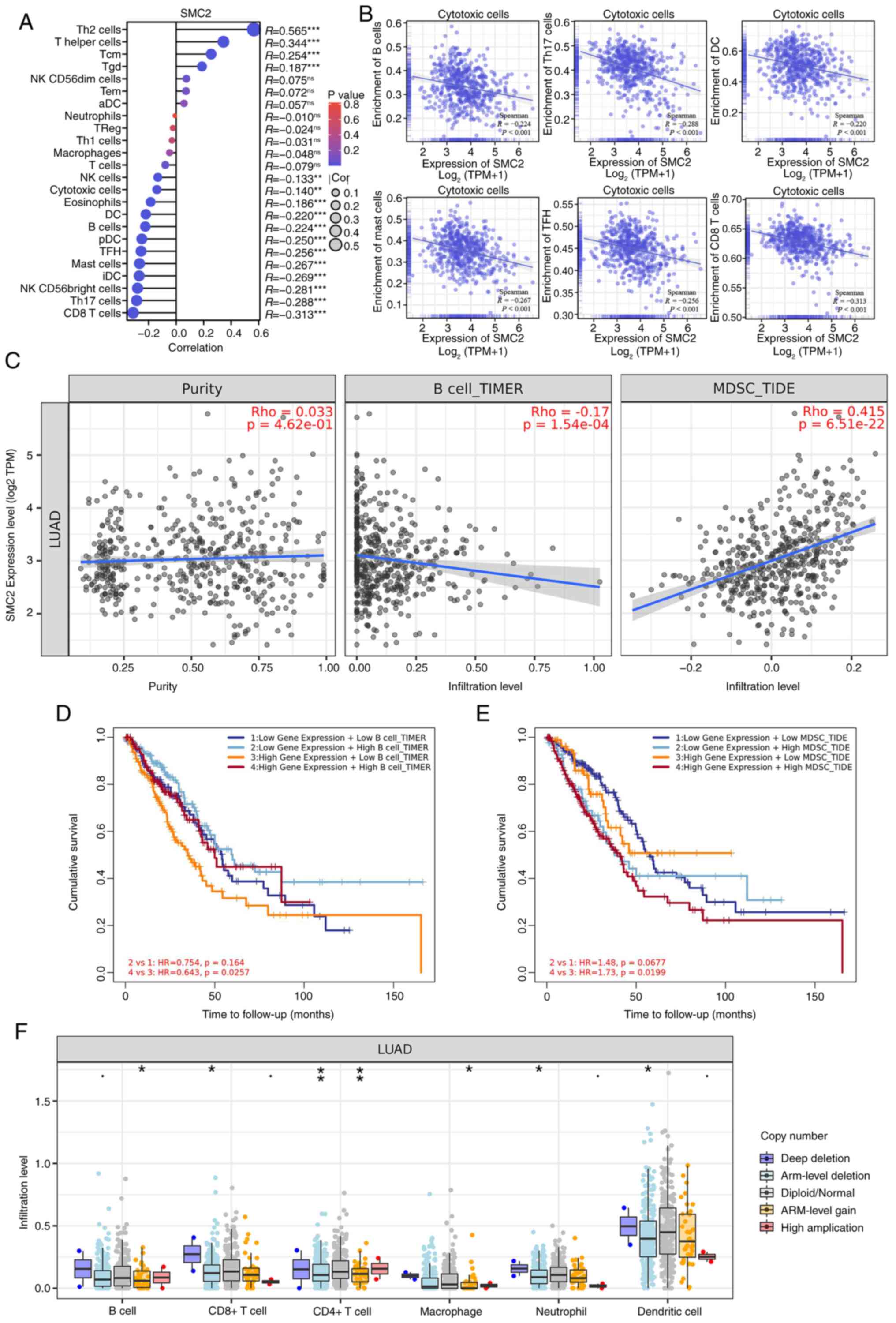
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018.PubMed/NCBI View Article : Google Scholar
|
|
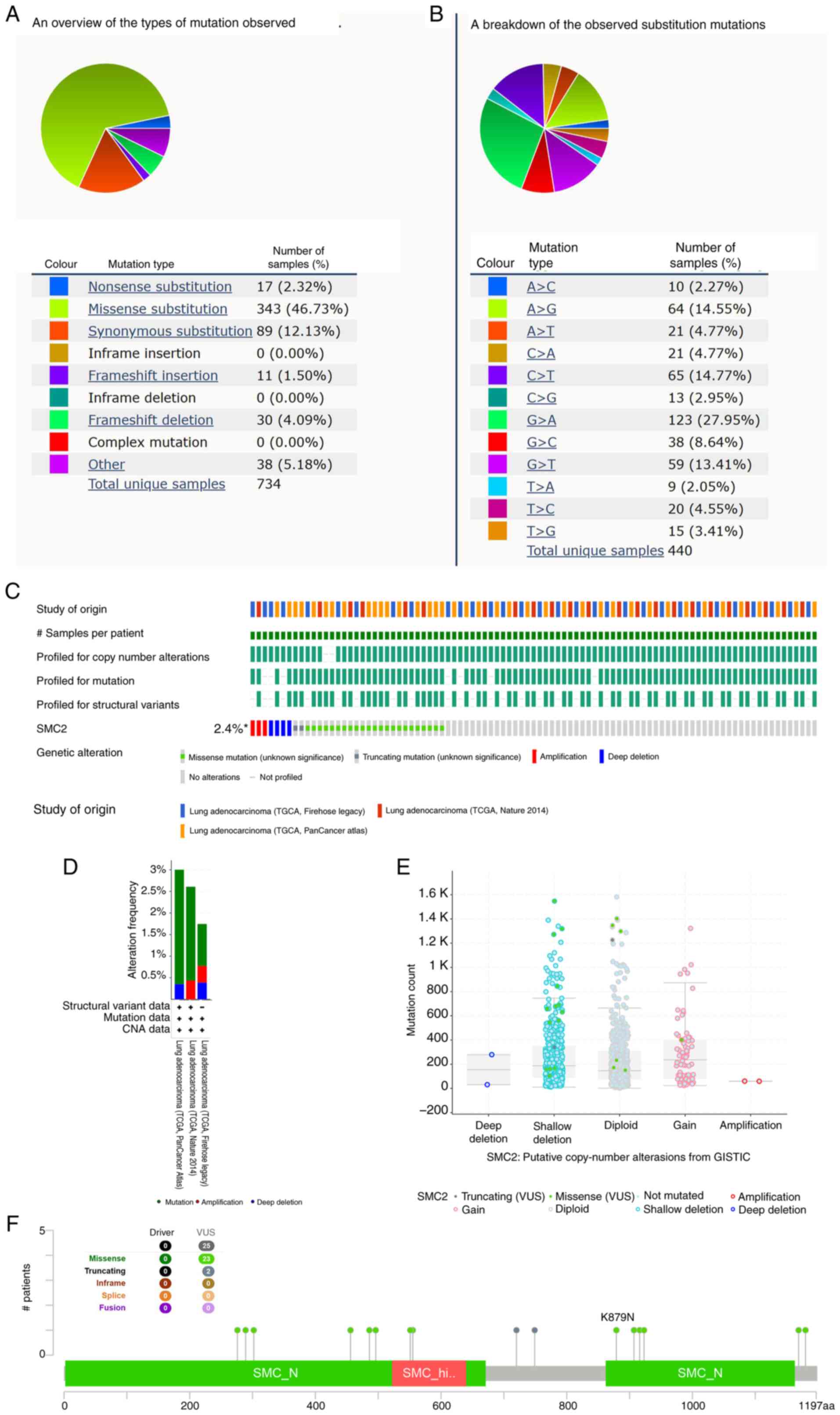
5
|
Hirano T: At the heart of the chromosome:
SMC proteins in action. Nat Rev Mol Cell Biol. 7:311–322.
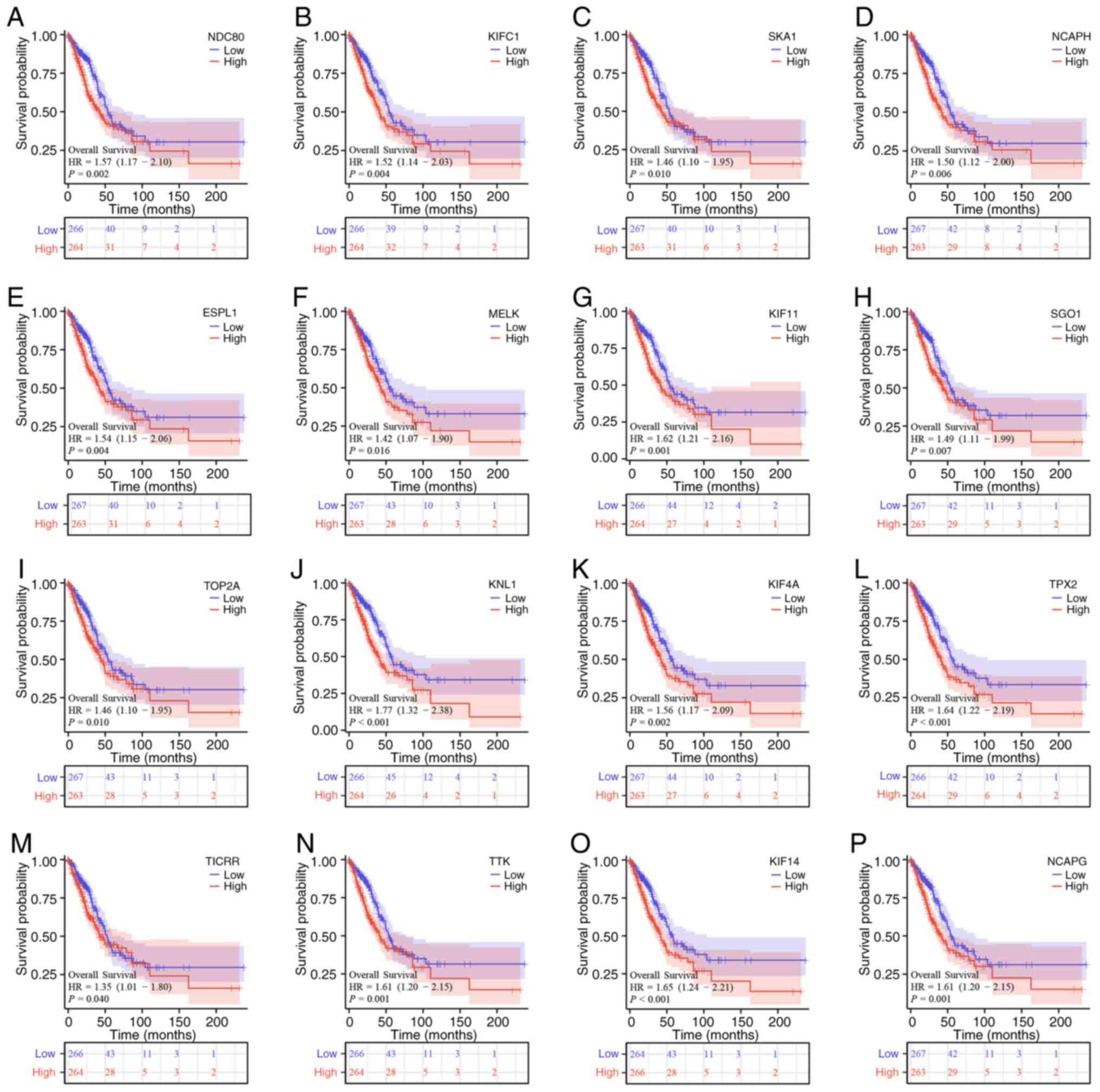
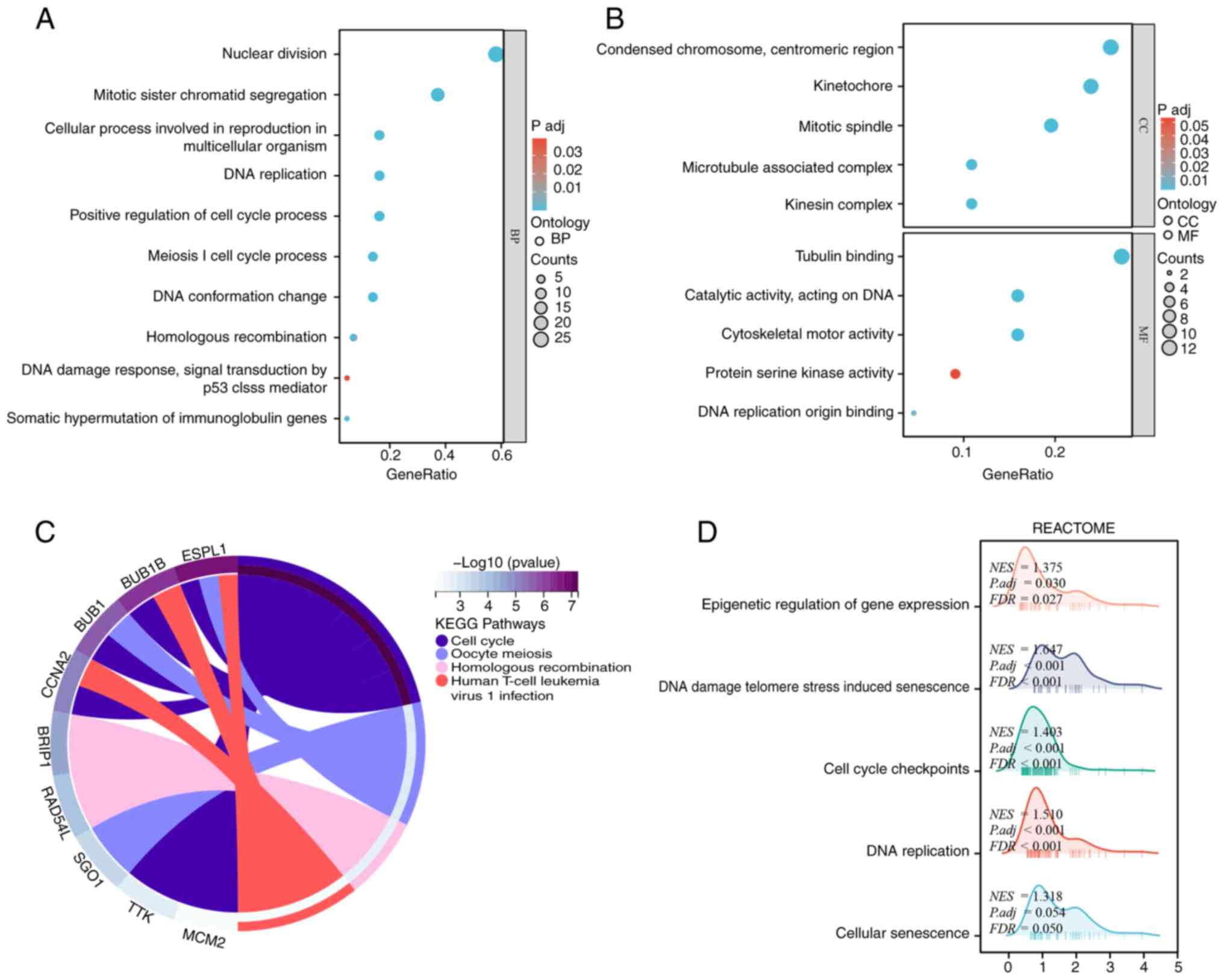
2006.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Zhao H, Shu L, Qin S, Lyu F, Liu F, Lin E,
Xia S, Wang B, Wang M, Shan F, et al: Extensive mutual influences
of SMC complexes shape 3D genome folding. Nature. 640:543–553.
2025.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dávalos V, Súarez-López L, Castaño J,
Messent A, Abasolo I, Fernandez Y, Guerra-Moreno A, Espín E,
Armengol M, Musulen E, et al: Human SMC2 protein, a core subunit of
human condensin complex, is a novel transcriptional target of the
WNT signaling pathway and a new therapeutic target. J Biol Chem.
287:43472–43481. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hudson DF, Marshall KM and Earnshaw WC:
Condensin: Architect of mitotic chromosomes. Chromosome Res.
17:131–144. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Paliulis LV and Nicklas RB:
Micromanipulation of chromosomes reveals that cohesion release
during cell division is gradual and does not require tension. Curr
Biol. 14:2124–2129. 2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Murakami-Tonami Y, Kishida S, Takeuchi I,
Katou Y, Maris JM, Ichikawa H, Kondo Y, Sekido Y, Shirahige K,
Murakami H and Kadomatsu K: Inactivation of SMC2 shows a
synergistic lethal response in MYCN-amplified neuroblastoma cells.
Cell Cycle. 13:1115–1131. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
12
|
Yadav S, Kowolik CM, Lin M, Zuro D, Hui
SK, Riggs AD and Horne DA: SMC1A is associated with radioresistance
in prostate cancer and acts by regulating epithelial-mesenchymal
transition and cancer stem-like properties. Mol Carcinog.
58:113–125. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Kraft B, Lombard J, Kirsch M, Wuchter P,
Bugert P, Hielscher T, Blank N and Krämer A: SMC3 protein levels
impact on karyotype and outcome in acute myeloid leukemia.
Leukemia. 33:795–799. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jiang L, Zhou J, Zhong D, Zhou Y, Zhang W,
Wu W, Zhao Z, Wang W, Xu W, He L, et al: Overexpression of SMC4
activates TGFβ/Smad signaling and promotes aggressive phenotype in
glioma cells. Oncogenesis. 6(e301)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
GTEx Consortium. Human genomics. The
genotype-tissue expression (GTEx) pilot analysis: Multitissue gene
regulation in humans. Science. 348:648–660. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Asplund A, Edqvist PH, Schwenk JM and
Pontén F: Antibodies for profiling the human proteome-The human
protein atlas as a resource for cancer research. Proteomics.
12:2067–2077. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49:D605–D612. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Canzler S and Hackermüller J: multiGSEA: A
GSEA-based pathway enrichment analysis for multi-omics data. BMC
Bioinformatics. 21(561)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Bamford S, Dawson E, Forbes S, Clements J,
Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR
and Wooster R: The COSMIC (Catalogue of Somatic Mutations in
Cancer) database and website. Br J Cancer. 91:355–358.
2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6(pl1)2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ru B, Wong CN, Tong Y, Zhong JY, Zhong
SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, et al: TISIDB: An
integrated repository portal for tumor-immune system interactions.
Bioinformatics. 35:4200–4202. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chi A, He X, Hou L, Nguyen NP, Zhu G,
Cameron RB and Lee JM: Classification of non-small cell lung
cancer's tumor immune micro-environment and strategies to augment
its response to immune checkpoint blockade. Cancers (Basel).
13(2924)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li J, Jie HB, Lei Y, Gildener-Leapman N,
Trivedi S, Green T, Kane LP and Ferris RL: PD-1/SHP-2 inhibits
Tc1/Th1 phenotypic responses and the activation of T cells in the
tumor microenvironment. Cancer Res. 75:508–518. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Feng Y, Liu H, Duan B, Liu Z, Abbruzzese
J, Walsh KM, Zhang X and Wei Q: Potential functional variants in
SMC2 and TP53 in the AURORA pathway genes and risk of pancreatic
cancer. Carcinogenesis. 40:521–528. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yan W, Wang DD, Zhang HD, Huang J, Hou JC,
Yang SJ, Zhang J, Lu L and Zhang Q: Expression profile and
prognostic values of SMC family members in HCC. Medicine
(Baltimore). 101(e31336)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Nie H, Wang Y, Yang X, Liao Z, He X, Zhou
J and Ou C: Clinical significance and integrative analysis of the
SMC family in hepatocellular carcinoma. Front Med (Lausanne).
8(727965)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Je EM, Yoo NJ and Lee SH: Mutational and
expressional analysis of SMC2 gene in gastric and colorectal
cancers with microsatellite instability. APMIS. 122:499–504.
2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Xu Y, Wang S, Xu B, Lin H, Zhan N, Ren J,
Song W, Han R, Cheng L, Zhang M and Zhang X: AURKA, TOP2A and MELK
are the key genes identified by WGCNA for the pathogenesis of lung
adenocarcinoma. Oncol Lett. 25(238)2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li C, Meng J and Zhang T: NCAPH is a
prognostic biomarker and associated with immune infiltrates in lung
adenocarcinoma. Sci Rep. 12(9578)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chen C, Guo Q, Song Y, Xu G and Liu L:
SKA1/2/3 serves as a biomarker for poor prognosis in human lung
adenocarcinoma. Transl Lung Cancer Res. 9:218–231. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhou F, Wang M, Aibaidula M, Zhang Z,
Aihemaiti A, Aili R, Chen H, Dong S, Wei W and Maimaitiaili A: TPX2
promotes metastasis and serves as a marker of poor prognosis in
non-small cell lung cancer. Med Sci Monit.
26(e925147)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ricciuti B, Arbour KC, Lin JJ, Vajdi A,
Vokes N, Hong L, Zhang J, Tolstorukov MY, Li YY, Spurr LF, et al:
Diminished efficacy of programmed death-(Ligand)1 inhibition in
STK11- and KEAP1-mutant lung adenocarcinoma is affected by KRAS
mutation status. J Thorac Oncol. 17:399–410. 2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wohlhieter CA, Richards AL, Uddin F,
Hulton CH, Quintanal-Villalonga À, Martin A, de Stanchina E, Bhanot
U, Asher M, Shah NS, et al: Concurrent mutations in STK11 and KEAP1
promote ferroptosis protection and SCD1 dependence in lung cancer.
Cell Rep. 33(108444)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Thadani R, Kamenz J, Heeger S, Muñoz S and
Uhlmann F: Cell-Cycle regulation of dynamic chromosome association
of the condensin complex. Cell Rep. 23:2308–2317. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang M, Chang M, Li C, Chen Q, Hou Z, Xing
B and Lin J: Tumor-microenvironment-activated reactive oxygen
species amplifier for enzymatic cascade cancer
starvation/chemodynamic/immunotherapy. Adv Mater.
34(e2106010)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Slack FJ and Chinnaiyan AM: The role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shao MM, Zhai K, Huang ZY, Yi FS, Zheng
SC, Liu YL, Qiao X, Chen QY, Wang Z and Shi HZ: Characterization of
the alternative splicing landscape in lung adenocarcinoma reveals
novel prognosis signature associated with B cells. PLoS One.
18(e0279018)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
van der Leun AM, Thommen DS and Schumacher
TN: CD8(+) T cell states in human cancer: Insights from single-cell
analysis. Nat Rev Cancer. 20:218–232. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang Y, Li Y, Jiang X, Gu Y, Zheng H, Wang
X, Zhang H, Wu J and Cheng Y: OPA1 supports mitochondrial
dynamics and immune evasion to CD8(+) T cell in lung
adenocarcinoma. PeerJ. 10(e14543)2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Guo Z, Liang H, Xu Y, Liu L, Ren X, Zhang
S, Wei S and Xu P: The role of circulating T Follicular helper
cells and regulatory cells in non-small cell lung cancer patients.
Scand J Immunol. 86:107–112. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Kalathil SG and Thanavala Y: Importance of
myeloid derived suppressor cells in cancer from a biomarker
perspective. Cell Immunol. 361(104280)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Islam SA and Luster AD: T cell homing to
epithelial barriers in allergic disease. Nat Med. 18:705–715.
2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zlotnik A and Yoshie O: The chemokine
superfamily revisited. Immunity. 36:705–716. 2012.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Matsukawa A, Hogaboam CM, Lukacs NW,
Lincoln PM, Evanoff HL and Kunkel SL: Pivotal role of the CC
chemokine, macrophage-derived chemokine, in the innate immune
response. J Immunol. 164:5362–5368. 2000.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Xing J, Zhang C, Yang X, Wang S, Wang Z,
Li X and Yu E: CXCR5+CD8+ T cells infiltrate
the colorectal tumors and nearby lymph nodes, and are associated
with enhanced IgG response in B cells. Exp Cell Res. 356:57–63.
2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ness TL, Ewing JL, Hogaboam CM and Kunkel
SL: CCR4 is a key modulator of innate immune responses. J Immunol.
177:7531–7539. 2006.PubMed/NCBI View Article : Google Scholar
|