Association of the expression of 5‑FU biomarkers with aging and prognosis in elderly patients with lung cancer treated with S‑1 adjuvant chemotherapy: Follow‑up results of the Setouchi Lung Cancer Group Study 1201
- Authors:
- Published online on: July 3, 2025 https://doi.org/10.3892/mco.2025.2874
- Article Number: 79
-
Copyright: © Soh et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Lung cancer is the leading cause of cancer-related death worldwide (1). Radical resection is the standard treatment for patients with clinical stage I to III non-small cell lung cancer (NSCLC). In Japan, adjuvant chemotherapy is recommended for patients with NSCLC having a pathological maximum tumor size with diameter of ≥2 cm, even with complete tumor resection. Elderly individuals tend to have poor treatment compliance because of physical and cognitive declines. The European Organization for Research and Treatment of Cancer and the International Society of Geriatric Oncology recommend adjuvant chemotherapy treatment for elderly individuals with NSCLC as postoperative adjuvant chemotherapy treatment can increase survival and should not be denied to patients based on age (2). Furthermore, prolonged exposure to some external factors and a decrease in DNA repair functions in elderly individuals may lead to various genetic abnormalities, suggesting that molecular mechanisms underlying carcinogenesis may differ by age group and may affect the clinical outcome of systemic therapy in geriatric patients with NSCLC.
S-1 (Taiho Pharmaceutical Co., Ltd, Tokyo, Japan) is an oral fluoropyrimidine agent, consisting of tegafur [a 5-fluorouracil (5-FU)prodrug] gimeracil [a dihydropyrimidine dehydrogenase (DPD) inhibitor that degrades 5-FU], and oteracil, a phosphorylation inhibitor. S-1 reduces the toxic effects of 5-FU in the gastrointestinal tract (3). S-1 monotherapy is effective in elderly patients with advanced NSCLC, and is an alternative treatment for platinum-doublet chemotherapy, has been indicated (4-6). Furthermore, the phase 2 clinical trial conducted by Setouchi Lung Cancer Group (SLCG1201) showed that the alternate-day and the daily oral administrations of S-1 for 14 consecutive days followed by 7-day rest were feasible in elderly patients with completely resected NSCLC (7).
5-FU sensitivity is influenced by its biomarkers, such as the target molecule, thymidylate synthase (TS) (8), fluoropyrimidine metabolizing enzymes (orotate phosphoribosyl transferase [OPRT] and thymidine phosphatase [TP] (9,10), and the 5-FU-degrading enzyme, DPD (11). Activating mutations in the epidermal growth factor receptor (EGFR) gene affects the efficacy of EGFR tyrosine kinase inhibitors (EGFR-TKIs). In addition, clinical and experimental studies have shown that EGFR mutations reduce the efficacy of adjuvant chemotherapy with oral uracil-tegafur (UFT) (Taiho Pharmaceutical Co., Ltd) (12,13). Excision repair cross-complementation group 1 (ERCC1) is implicated in DNA repair pathways. Further, ERCC1 expression is associated with poor prognosis in patients with NSCLC treated with cisplatin-based chemotherapy, and low ERCC1 expression correlated with better prognosis in patients with gastric and colon cancer treated with 5-FU (14). However, the relationship between the expression levels of ERCC1 and the therapeutic efficacy of 5-FU chemotherapy in patients with NSCLC remains unclear.
This study investigated the effect of aging on the expression levels of 5-FU biomarkers by analyzing RNA-seq data from The Cancer Genome Atlas (TCGA) database and assessed the ability of these biomarkers to predict the clinical outcomes of elderly patients with NSCLC who underwent complete resection in the SCLG1201 followed by adjuvant chemotherapy with S-1.
Materials and methods
Analysis of TCGA datasets
Data on RNA-Seq gene expression profiles of TCGA samples were obtained from the Genomic Data Commons Portal (https://portal.gdc.cancer.gov). The datasets TCGA-LUAD and TCGA-LUSC were downloaded in March 2023. The accession numbers of the data are luad_tcga_pan_can_atlas_2018 and lusc_tcga_pan_can_atlas_2018, respectively. Data on the z-scores of six genes (DPYD, TYMP, TYMS, UMPS, ERCC1, and EGFR), EGFR mutational status, and demographic and clinicopathological characteristics (age, sex, race, and histology) were retrieved and analyzed.
Patients' selection, ethics approval and consent to participate
We enrolled patients who had agreed to participate in both the SLCG1201 (University Hospital Medical Information Network ID: UMIN000007819) (7) and this follow-up study. Patients provided written informed consents for both studies. The SCLG1201 was approved by Okayama University Certified Review Board (Approval number: CRB18-011), followed by the confirmation of each participating institution.
The follow-up study was approved by the Ethics Committee of the Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences and Okayama University Hospital (Okayama, Japan) (Approval number: Rin1610, later revised as Rin1945) and by the institutional review boards of the participating institutions. The study's data was managed by the Epidemiological and Clinical Research Information Network (Kyoto, Japan), a non-profit organization.
Sample collection and preparation
Surgical specimens were fixed in 10% formaldehyde and embedded in paraffin. Formaldehyde-fixed and paraffin-embedded (FFPE) tumor blocks were sectioned and overlaid on non-coating slides. Two slides received 5-µm-thick sections, and five received 10-µm-thick sections at each participating institution. The slides were immediately sent to the Department of Thoracic Surgery of Okayama University Hospital. After confirmation of the quality of each section and anonymization, the slides were sent to FALCO Biosystems Ltd. (Kyoto, Japan).
Representative hematoxylin and eosin-stained slides were prepared from the 5-µm-thick FFPE slides and reviewed by a pathologist for the manual macrodissection of tumor tissues. Tumor tissues from the 10-µm-thick FFPE slides were dissected using a scalpel. RNA was isolated from dissected tumor tissues using the RNeasy FFPE Kit (Qiagen, Chatsworth, GA, USA) and reverse transcribed using the High-Capacity Reverse Transcription Kit (Life Technologies, Foster City, CA, USA) according to the manufacturer's instruction.
Quantitative reverse-transcription PCR
The expression levels of six genes (TP, TS, DPD, OPRT, ERCC1, and EGFR) were quantitated using TaqMan real-time PCR (TaqMan array card; Life Technologies). Briefly, 2.5 µl of cDNA was pre-amplified using TaqMan PreAmp Master Mix (2x) (Life Technologies) and a pool of TaqMan® Gene Expression Assays (0.2x) in a 10-µl PCR reaction volume. The pre-amplification cycling conditions were as follows: one cycle at 95˚C for 10 min, followed by 14 cycles at 95˚C for 15 sec and at 60˚C for 4 min. Amplified cDNA samples were diluted 20 times in TE buffer, and 25 µl of a cDNA sample was added to 25 µl of RNase-free water and 50 µl of 2x TaqMan Gene Expression Master Mix (Life Technologies). The mixture was transferred to a loading port of the TaqMan low-density array microfluidics card. The array card was centrifuged twice and sealed. PCR amplification was performed using the Applied Biosystems Prism 7900HT Sequence Detection System (Life Technologies) under the following thermal cycling conditions: one cycle at 50.0˚C for 2 min and 94.5˚C for 10 min, followed by 40 cycles at 97.0˚C for 30 sec and 59.7˚C for 1 min. β-actin (ACTB) served as the housekeeping gene. The assay IDs used in the array card are shown in Table SI. The cycle threshold (Ct) value, inversely proportional to the amount of cDNA, was calculated. The relative mRNA expression levels were expressed as the ratios (differences between Ct values) between the gene of interest and the reference gene.
Statistical analysis
Categorized variables were compared using the chi-square test or Fisher's exact test, and continuous variables were compared using the Mann-Whitney U test. The correlation among multiple continuous variables was assessed using Pearson's correlation coefficient. Follow-up data were obtained from the SLCG1201(7). Overall survival (OS) and recurrence-free survival (RFS) were analyzed using the Kaplan-Meier method with the log-rank test and univariate and multivariate Cox proportional hazards regression. P<0.05 was defined as the threshold for statistical significance. All statistical analyses were performed using JMP version 9.0.2 Program for Windows (SAS Institute Inc., Cary, NC, USA) and GraphPad Prism 9 (GraphPad Software, La Jolla, CA, USA).
Results
Impact of age on gene expression in silico
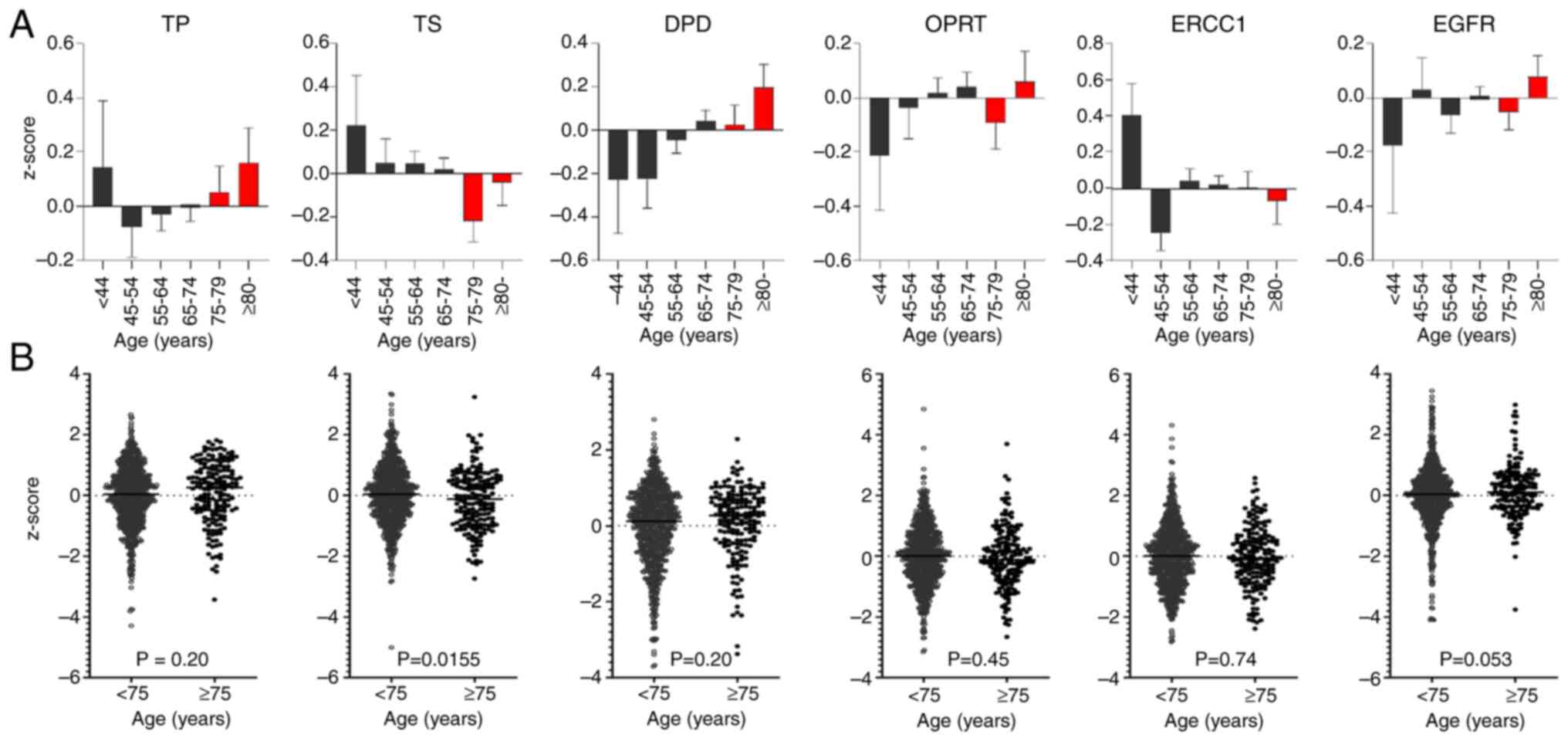
We investigated the impact of age on the expression of six genes by analyzing RNA-seq data of 955 samples (776 samples from patients younger than 75 years and 179 from patients aged ≥75 years) from the TCGA database (Tables SII and SIII). Gene expression levels were compared across age groups (Fig. 1). Using the Mann-Whitney U test, TS expression decreased significantly with age, especially in the age group ≥75 years (P=0.0155). In contrast, DPD and TP expression levels increased to a similar extent in the age groups <75 and ≥75 years.
Patient characteristics
The SLCG1201 study enrolled 101 patients from 19 institutions in Japan between May 2012 to April 2016, with 97 patients receiving the allocated intervention (7). The inclusion and exclusion criteria have been previously described (7). ‘Never smoked’ was defined as <100 cigarettes in the patient's life and ‘ever smoked’ was defined as the patient having ≥100 cigarettes in their life. Among them, 90 agreed to participate in this study between May 2013 to April 2016. One sample was excluded from the analysis because of the absence of tumors in the slides. Thus, 89 patients were included in the study. The baseline characteristics of the cohort are shown in Table I. The median age was 77 years (range, 75-87 years), and 65 patients (73.0%) were men. Fifty-seven (64.0%) patients had adenocarcinoma histology and 54 (60.7%) were pathological stage (pStage) IA (T1bN0M0)/IB. EGFR mutations were present in 27 (30.3%) patients.
The 17 institutions that participated in the SLCG1201 follow-up study, listed in order based on the number of registered patients, are as follows: Kurashiki Central Hospital, Fukushima Medical University Hospital, Okayama University Hospital, Kawasaki Medical School Hospital, Hiroshima City Hiroshima Citizens Hospital, Chugoku Central Hospital, Japanese Red Cross Nagasaki Genbaku Hospital, National Hospital Organization Nagara Medical Center, Okayama Saiseikai General Hospital, Saga-Ken Medical Center Koseikan, Kyoto University Hospital, Okayama Rosai Hospital, National Hospital Organization Iwakuni Clinical Center, Shimane Prefectural Central Hospital, National Hospital Organization Yamaguchi-Ube Medical Center, Tottori University Hospital, National Hospital Organization Kure Medical Center and Chugoku Cancer Center.
Relative gene expression and EGFR mutations
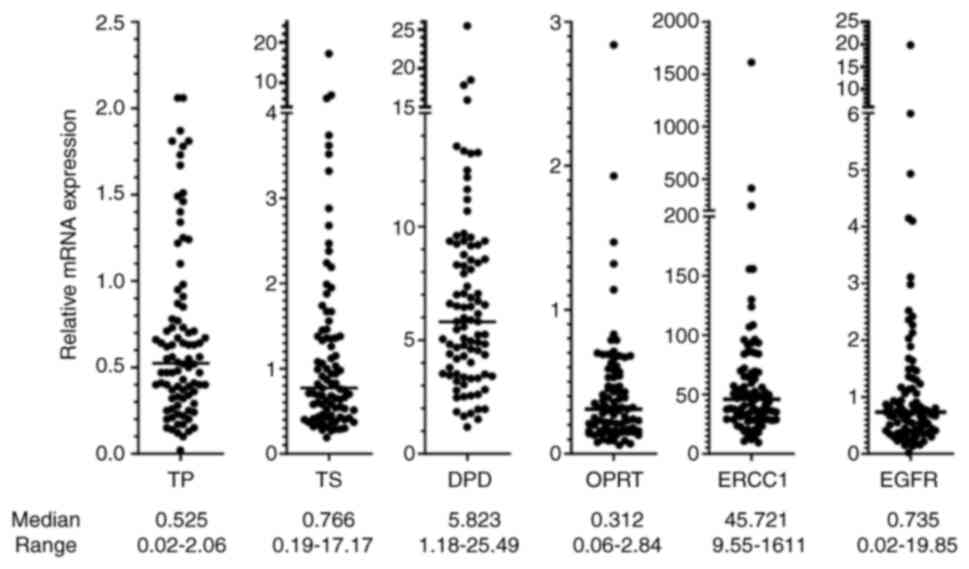
The relative mRNA expression levels of six genes are shown in Fig. 2. The median Ct value of ACTB of all 89 samples was 15.69 (range, 11.65-26.66), indicating that the mRNA quality was adequate. However, some genes were not amplified in three samples: TP and EGFR in one sample (the Ct of ACTB was 26.66) and OPRT in two samples (the Ct of ACTB was 21.72 and 19.88, respectively). There were no significant correlations between the relative mRNA expression levels of these genes (Fig. S1).
The associations of clinicopathological factors with expression profiles are shown in Table II. EGFR mutations were significantly frequent in females, never smokers, patients with poor performance status (PS), and patients with adenocarcinoma, consistent with previous studies (15-17). TS and OPRT were upregulated in males, and DPD and EGFR were upregulated in females. TS was upregulated in ever smokers, and DPD and EGFR were upregulated in never smokers. TS, OPRT and ERCC1 were upregulated in patients with non-adenocarcinoma histology, and DPD was upregulated in patients with adenocarcinoma histology.
EGFR mutations are important driver mutations in NSCLC; hence, we investigated the relationship between the expression levels of six genes and EGFR mutations. EGFR mutations were significantly associated with the upregulation of DPD (P=0.0066) and EGFR (P<0.0001) and the downregulation of TS (P=0.0125) and ERCC1 (P=0.0015) (Table II).
Prognostic impact of clinicopathological factors and molecular profiles
The patients were divided into two groups based on the cutoff values of the median mRNA expression of six genes.
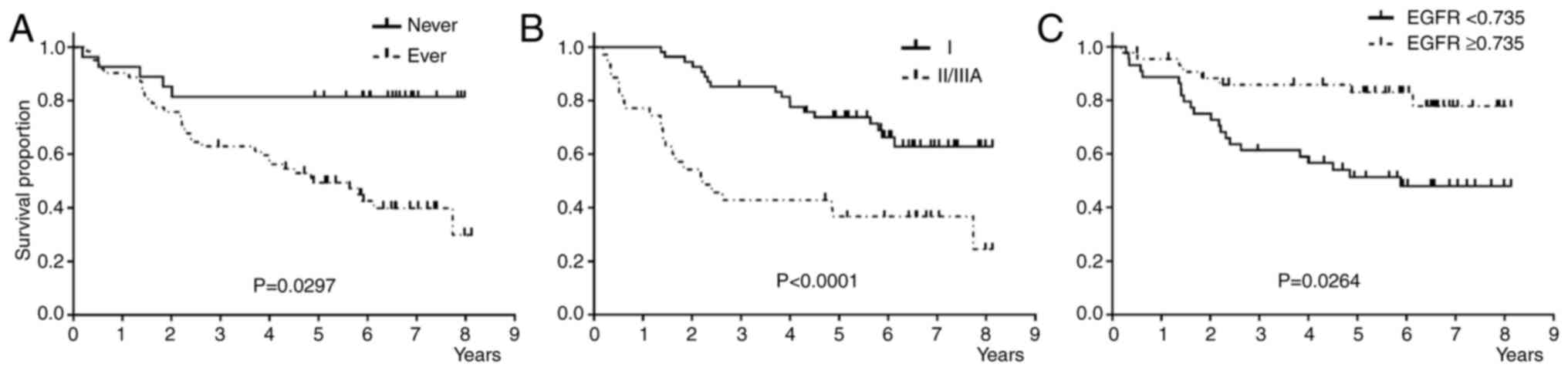
Univariate analysis showed that never smokers, patients with pStage I, and patients with EGFR upregulation showed significantly better RFS than the groups (Fig. 3, and Table IIIA). Multivariate analysis, including the factors with P-value ≤0.1, showed that pStage I was an independent favorable prognostic factor for RFS (Table 3A). Stepwise multivariate analysis showed that pStage I and never smoking status were independent favorable prognostic factors for RFS (Table SIVA).
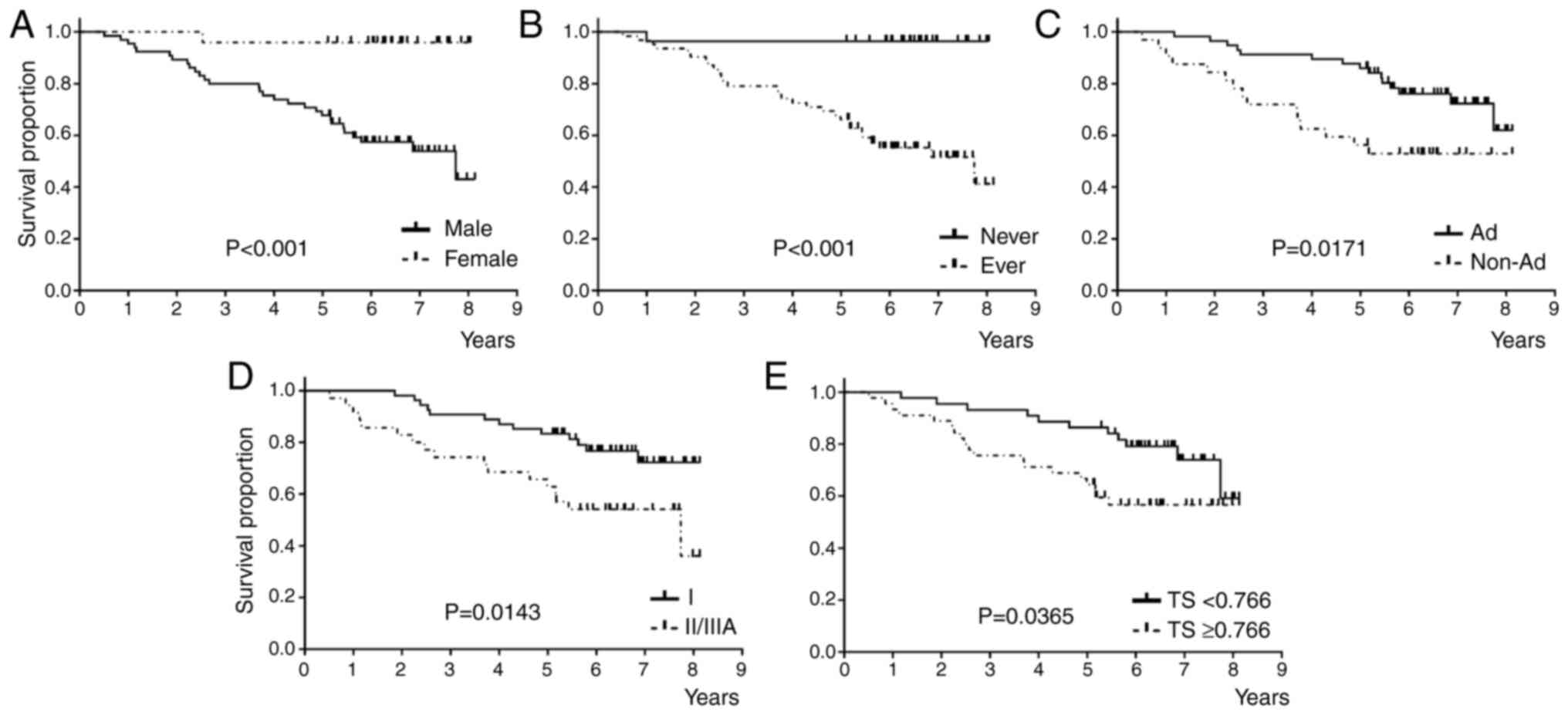
Univariate analysis revealed that female sex, never smoking status, adenocarcinoma histology, pStage I, and TS downregulation were significantly associated with better OS than the others (Fig. 4 and Table IIIB). Multivariate analysis, including the factors with P-value ≤0.1, showed that pStage I was an independent favorable prognostic factor for OS (Table IIIB). Stepwise multivariate analysis indicated that pStage I and never smoking status were independent favorable prognostic factors for OS (Table SIVB).
Discussion
Aging affects gene expression via mRNA splicing and genetic regulation (18,19). The expression of several genes is affected by age in multiple tissues (20,21). To our knowledge, the six genes investigated in this study have not been previously linked with aging. However, the analysis of TCGA RNA-Seq data suggests that TS expression is affected by age. Aging exacerbates the effects of folate deficiency, including its downstream pathway (22). TS is a key downstream molecular target of folate and plays a crucial role in DNA replication and repair. TS expression is tightly regulated by epigenetic modifications (e.g., DNA methylation), transcription factors (E2F, p53, c-Myc), microRNAs, folate availability, hypoxia, and drug interactions. Genome-wide DNA methylation declines with age (22), which may contribute to the downregulation of TS. Additionally, the age-related downregulation of TS expression may be due to epigenetic repression, reduced cell proliferation, metabolic changes, chronic inflammation, and oxidative stress. These findings support the notion that TS expression is influenced by age and that the expression levels of 5-FU biomarkers changes with age, potentially impacting the clinical outcomes of adjuvant treatment in elderly patients with NSCLC compared with younger patients.
This study examined the mRNA expression of six biomarker genes associated with 5-FU chemotherapy in patients aged ≥75 years with pStage IA (2 cm) to IIIA NSCLC. We investigated the association of these markers with clinicopathological characteristics and prognosis. The upregulation of EGFR and DPD genes and downregulation of ERCC1 and TS genes were significantly associated with the presence of EGFR mutation. The univariate analysis for RFS and OS revealed that the downregulation of EGFR was an unfavorable factor for RFS, and the upregulation of TS was an unfavorable factor for OS. Multivariate analysis showed that pStage II/III was identified as an independent unfavorable factor for both RFS and OS.
EGFR mutations were significantly associated with the upregulation of EGFR and DPD and the downregulation of ERCC1 and TS. Consistent with these findings, previous studies have shown that mutant alleles specific imbalance is common in mutant EGFR cells and correlates with increased mutant allele transcription (23-25). The upregulation of DPD is associated with EGFR mutations in clinical samples and cell lines (26). Moreover, EGFR signaling regulates DPD expression via Sp-1 in EGFR-mutant cells (27). These findings support the notion that EGFR mutations contribute to resistance to 5-FU-based therapies by upregulating DPD. Regarding ERCC1, an experimental study showed that increased DNA damage and reduced damage repair (ERCC1 and RAD51 foci formation) were more common in EGFR exon 19 deletion mutant cells than in EGFR wild-type cells (28). Furthermore, ERCC1 expression was increased by inhibiting EGFR exon 19 deletion signals and decreased by blocking wild-type EGFR signals. NSCLC specimens with EGFR activating mutations tend to have low ERCC1 mRNA levels (28-31). ERCC1 and TS are essential for DNA synthesis and repair, and TS expression was decreased in EGFR-mutant lung cancer specimens, consistent with our findings (29,31).
Univariate analysis showed that EGFR downregulation and TS upregulation were significantly associated with unfavorable RFS and OS, respectively. To our knowledge, the prognostic impact of EGFR mRNA expression has not been well-investigated in patients who received 5-FU adjuvant chemotherapy after complete resection. However, we have previously showed that adjuvant UFT improved prognosis in patients without EGFR mutations but not in patients with EGFR mutation (12), in line with a previous study (13). In contrast, a meta-analysis of 18 studies involving 2972 patients conducted before EGFR-TKIs were developed showed that EGFR overexpression was not associated with favorable prognosis (combined HR of 1.14 with 95%CI of 0.97 to 1.34; p=0.103) (32). Although our univariate analysis indicated that EGFR upregulation was associated with favorable RFS, its impact on RFS in patients receiving 5-FU-based therapies remains controversial. Several studies and meta-analyses showed that low TS protein expression was significantly associated with favorable prognosis in patients with lung cancer who received S-1-based chemotherapy (33,34) and pemetrexed-based chemotherapy (35). Furthermore, TS overexpression was associated with poor prognosis in patients treated with 5-FU and UFT, including those with NSCLC (33,36,37) and gastrointestinal cancers (38). Moreover, in vitro studies have suggested that EGFR-TKIs induce TS downregulation in TKI-resistant NSCLC cells with MET amplification but not in cells harboring the T790M EGFR mutation (39-41). These findings suggest a link between TS expression and EGFR mutations.
Regarding EGFR mutation subtypes, a pooled analysis of 12 clinical trials involving patients with advanced NSCLC and EGFR mutations indicated that exon 19 deletion was significantly associated with better clinical outcomes than the L858R mutation after EGFR-TKI treatment (42). Additionally, preclinical studies have demonstrated that the molecular differences in these two subtypes influence the efficacy of EGFR-TKIs (43-46). Therefore, we compared the expression levels of six genes corresponding to different EGFR mutation subtypes. There were no significant differences in expression because of the small number of cases with specific EGFR mutation subtypes (12 cases with exon 19 deletions, 9 cases with L858R mutation, and 3 cases with rare mutations) (data not shown). Exon 19 deletion was significantly more prevalent in younger patients than the L858R mutation (42), supporting age affects the mechanisms of lung cancer. Although S-1 monotherapy was effective and feasible as a subsequent-line treatment in a small cohort of elderly patients treated with anti-cancer therapies, including EGFR-TKIs (47), large-scale studies are needed to evaluate the efficacy of S-1 chemotherapy, including its role as adjuvant therapy, in elderly patients with EGFR-mutated NSCLC, with a focus on EGFR mutation subtypes.
This study has some limitations. First, the small sample size may limit the generalizability of our findings to larger populations. Thus, studies with larger, independent and diverse cohorts are needed to confirm the robustness and reproducibility of our results. Notwithstanding, all patients were monitored until death or for at least 5 years from the registration. Second, as a preliminary study, we performed receiver operating characteristic (ROC) analysis using survival outcomes (dead or alive). However, except for DPD [which had an area under the curve (AUC) of 0.631 and a P-value of 0.0332], the AUC values for the other markers were below 0.543, with P-values exceeding 0.10. We also conducted ROC analysis based on recurrence events; however, this approach similarly failed to yield suitable cutoff values (data not shown). Additionally, we observed a substantial imbalance in the number of subjects classified according to the ROC-derived cutoffs. Given these limitations, we determined the cutoff values using median values instead of ROC analysis, as the small number of patients did not allow for the determination of suitable cutoff values for mRNA expression. Third, although mRNA was detected using a commercial real-time PCR assay using primers for a specific lesion of the target gene, we did not assess the correlation between mRNA expression level and protein expression levels. Nevertheless, as a preliminary study, we evaluated the correlation between mRNA and protein expression levels for TP, DPD, and EGFR (data not shown). Experimental and clinical studies have shown that the non-synonymous SNP 538G>A in MRP8/ABCC11, an ABC transporters for which 5-FU, methotrexate, and pemetrexed are substrates, is a potential biomarker for S-1 treatment (48,49). Clarifying the clinical implications of these candidate biomarkers is warranted. Fourth, we found that pStage was an independent prognostic factor but did not account for other potential confounding factors, such as comorbidities. Thus, future studies with more patient data and rigorous adjustments for confounders are needed to elucidate the prognostic value of 5-FU biomarkers.
In conclusion, the analysis of TCGA data showed that TS expression was significantly decreased with age. Univariate analysis indicated that, among 5-FU derivatives, TS downregulation and EGFR upregulation were favorable prognostic markers in elderly patients with NSCLC who underwent radical resection followed by adjuvant chemotherapy with S-1. Although pStage was an independent prognostic factor in the multivariable analysis, the findings suggested that elderly patients with NSCLC exhibited low TS expression, which might have improved the clinical outcomes of S-1 adjuvant chemotherapy in this population.
Supplementary Material
Correlation between relative mRNA expression levels of six genes. EGFR, epidermal growth factor receptor; DPD, dihydropyrimidine dehydrogenase; TP, thymidine phosphorylase; TS, thymidylate synthase; OPRT, orotate phosphoribosyl transferase; ERCC1, excision repair cross-complementation group 1.
Lists of assay IDs for Taqman Gene Expression Assays.
Downloaded data from The Cancer Genome Atlas.
Patient characteristics of 955 NSCLC samples downloaded from The Cancer Genome Atlas.
Multivariate analyses of prognosis using the stepwise method.
Acknowledgements
We thank Ms. Yumi Miyashita [Epidemiological and Clinical Research Information Network (a non-profit organization), Kyoto, Japan] for data management.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
JSo, HYa and SToK wrote the manuscript. JSo, HD and SToy were responsible for study conception and design. KH, HYo and SToy prepared the study protocol. HYa, NO, HS, MN, TFujiw, KG, IS, TFujin, MK, YTe, NF, KK, SK, MY, HI, MI, HN and YY collected the data. JSo, YTak, HT, HS, and STom performed the experiments. HYa, JSo, NO, SM, KM, JSa, HD and SToy analyzed and interpreted the data. All authors read and approved the final manuscript. JSo and HYa confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The SCLG1201 study was approved by the Okayama University Certified Review Board (approval no. CRB18-011), followed by the confirmation of each participating institution. The follow-up study was approved by the Ethics Committee, the Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences and Okayama University Hospital (Okayama, Japan) (approval no. Rin1945) and by the institutional review boards of the participating institutions. We enrolled patients who had agreed to participate in the SLCG1201 (University Hospital Medical Information Network ID: UMIN000007819) and this follow-up study. Patients provided written informed consent for both studies.
Patient consent for publication
Patients provided written informed consent for both studies for the publication of the findings.
Competing interests
The authors declare that they have no competing interests.
Use of artificial intelligence tools
During the preparation of this work, artificial intelligence tools were used only to improve the readability and language of a part of the manuscript, and subsequently, the authors revised and edited the content produced by the artificial intelligence tools as necessary, taking full responsibility for the ultimate content of the present manuscript. Then, we asked Editage (www.editage.jp) for final English language editing.
References
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Pallis AG, Gridelli C, Wedding U, Faivre-Finn C, Veronesi G, Jaklitsch M, Luciani A and O'Brien M: Management of elderly patients with NSCLC; updated expert's opinion paper: EORTC elderly task force, lung cancer group and international society for geriatric oncology. Ann Oncol. 25:1270–1283. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Shirasaka T, Shimamato Y, Ohshimo H, Yamaguchi M, Kato T, Yonekura K and Fukushima M: Development of a novel form of an oral 5-fluorouracil derivative (S-1) directed to the potentiation of the tumor selective cytotoxicity of 5-fluorouracil by two biochemical modulators. Anticancer Drugs. 7:548–557. 1996.PubMed/NCBI View Article : Google Scholar | |
|
Nishiyama O, Taniguchi H, Kondoh Y, Takada K, Baba K, Saito H, Sugino Y, Yamamoto M, Ogasawara T, Kondo M, et al: Phase II study of S-1 monotherapy as a first-line treatment for elderly patients with advanced nonsmall-cell lung cancer: The Central Japan lung study group trial 0404. Anticancer Drugs. 22:811–816. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Shiroyama T, Kijima T, Komuta K, Yamamoto S, Minami S, Ogata Y, Okafuji K, Imamura F, Hirashima T, Tachibana I, et al: Phase II tailored S-1 regimen study of first-line chemotherapy in elderly patients with advanced and recurrent non-small cell lung cancer. Cancer Chemother Pharmacol. 70:783–789. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Goto H, Okano Y, Machida H, Hatakeyama N, Ogushi F, Haku T, Kanematsu T, Urata T, Kakiuchi S, Hanibuchi M, et al: Phase II study of tailored S-1 monotherapy with a 1-week interval after a 2-week dosing period in elderly patients with advanced non-small cell lung cancer. Respir Investig. 56:80–86. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Yamamoto H, Soh J, Okumura N, Suzuki H, Nakata M, Fujiwara T, Gemba K, Sano I, Fujinaga T, Kataoka M, et al: Randomized phase II study of daily versus alternate-day administrations of S-1 for the elderly patients with completely resected pathological stage IA (tumor diameter >2 cm)-IIIA of non-small cell lung cancer: Setouchi lung cancer group study 1201. PLoS One. 18(e0285273)2023.PubMed/NCBI View Article : Google Scholar | |
|
Rustum YM, Harstrick A, Cao S, Vanhoefer U, Yin MB, Wilke H and Seeber S: Thymidylate synthase inhibitors in cancer therapy: Direct and indirect inhibitors. J Clin Oncol. 15:389–400. 1997.PubMed/NCBI View Article : Google Scholar | |
|
Peters GJ, Laurensse E, Leyva A, Lankelma J and Pinedo HM: Sensitivity of human, murine, and rat cells to 5-fluorouracil and 5'-deoxy-5-fluorouridine in relation to drug-metabolizing enzymes. Cancer Res. 46:20–28. 1986.PubMed/NCBI | |
|
Peters GJ, van Groeningen CJ, Laurensse EJ and Pinedo HM: A comparison of 5-fluorouracil metabolism in human colorectal cancer and colon mucosa. Cancer. 68:1903–1909. 1991.PubMed/NCBI View Article : Google Scholar | |
|
Beck A, Etienne MC, Chéradame S, Fischel JL, Formento P, Renée N and Milano G: A role for dihydropyrimidine dehydrogenase and thymidylate synthase in tumour sensitivity to fluorouracil. Eur J Cancer. 30A:1517–1522. 1994.PubMed/NCBI View Article : Google Scholar | |
|
Suehisa H, Toyooka S, Hotta K, Uchida A, Soh J, Fujiwara Y, Matsuo K, Ouchida M, Takata M, Kiura K and Date H: Epidermal growth factor receptor mutation status and adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. J Clin Oncol. 25:3952–3957. 2007.PubMed/NCBI View Article : Google Scholar | |
|
Tsutani Y, Ito M, Shimada Y, Ito H, Ikeda N, Nakayama H and Okada M: The impact of epidermal growth factor receptor mutation status on adjuvant chemotherapy for patients with high-risk stage I lung adenocarcinoma. J Thorac Cardiovasc Surg. 164:1306–1315.e4. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Wei J, Zou Z, Qian X, Ding Y, Xie L, Sanchez JJ, Zhao Y, Feng J, Ling Y, Liu Y, et al: ERCC1 mRNA levels and survival of advanced gastric cancer patients treated with a modified FOLFOX regimen. Br J Cancer. 98:1398–1402. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, et al: Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 97:339–346. 2005.PubMed/NCBI View Article : Google Scholar | |
|
Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, et al: The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 11:1167–1173. 2005.PubMed/NCBI | |
|
Soh J, Toyooka S, Matsuo K, Yamamoto H, Wistuba II, Lam S, Fong KM, Gazdar AF and Miyoshi S: Ethnicity affects EGFR and KRAS gene alterations of lung adenocarcinoma. Oncol Lett. 10:1775–1782. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Viñuela A, Snoek LB, Riksen JA and Kammenga JE: Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 20:929–937. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Li Z, Wright FA and Royland J: Age-dependent variability in gene expression in male Fischer 344 rat retina. Toxicol Sci. 107:281–292. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Yang J, Huang T, Petralia F, Long Q, Zhang B, Argmann C, Zhao Y, Mobbs CV, Schadt EE, Zhu J, et al: Synchronized age-related gene expression changes across multiple tissues in human and the link to complex diseases. Sci Rep. 5(15145)2015.PubMed/NCBI View Article : Google Scholar | |
|
Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K, et al: The transcriptional landscape of age in human peripheral blood. Nat Commun. 6(8570)2015.PubMed/NCBI View Article : Google Scholar | |
|
Jang H, Mason JB and Choi SW: Genetic and epigenetic interactions between folate and aging in carcinogenesis. J Nutr. 135 (Suppl):2967S–2971S. 2005.PubMed/NCBI View Article : Google Scholar | |
|
Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C, et al: Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 4(e7464)2009.PubMed/NCBI View Article : Google Scholar | |
|
Liang Z, Zhang J, Zeng X, Gao J, Wu S and Liu T: Relationship between EGFR expression, copy number and mutation in lung adenocarcinomas. BMC Cancer. 10(376)2010.PubMed/NCBI View Article : Google Scholar | |
|
Pinter F, Papay J, Almasi A, Sapi Z, Szabo E, Kanya M, Tamasi A, Jori B, Varkondi E, Moldvay J, et al: Epidermal growth factor receptor (EGFR) high gene copy number and activating mutations in lung adenocarcinomas are not consistently accompanied by positivity for EGFR protein by standard immunohistochemistry. J Mol Diagn. 10:160–168. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Mochinaga K, Tsuchiya T, Nagasaki T, Arai J, Tominaga T, Yamasaki N, Matsumoto K, Miyazaki T, Nanashima A, Hayashi T, et al: High expression of dihydropyrimidine dehydrogenase in lung adenocarcinoma is associated with mutations in epidermal growth factor receptor: Implications for the treatment of non-small-cell lung cancer using 5-fluorouracil. Clin Lung Cancer. 15:136–144.e4. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Tominaga T, Tsuchiya T, Mochinaga K, Arai J, Yamasaki N, Matsumoto K, Miyazaki T, Nagasaki T, Nanashima A, Tsukamoto K and Nagayasu T: Epidermal growth factor signals regulate dihydropyrimidine dehydrogenase expression in EGFR-mutated non-small-cell lung cancer. BMC Cancer. 16(354)2016.PubMed/NCBI View Article : Google Scholar | |
|
Zhang L, Pradhan B, Guo L, Meng F and Zhong D: EGFR exon 19-deletion aberrantly regulate ERCC1 expression that may partly impaired DNA damage repair ability in non-small cell lung cancer. Thorac Cancer. 11:277–285. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Lin CS, Liu TC, Lai JC, Yang SF and Tsao TC: Evaluating the prognostic value of ERCC1 and thymidylate synthase expression and the epidermal growth factor receptor mutation status in adenocarcinoma non-small-cell lung cancer. Int J Med Sci. 14:1410–1417. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Gandara DR, Grimminger P, Mack PC, Lara PN Jr, Li T, Danenberg PV and Danenberg KD: Association of epidermal growth factor receptor activating mutations with low ERCC1 gene expression in non-small cell lung cancer. J Thorac Oncol. 5:1933–1938. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Ren S, Chen X, Kuang P, Zheng L, Su C, Li J, Li B, Wang Y, Liu L, Hu Q, et al: Association of EGFR mutation or ALK rearrangement with expression of DNA repair and synthesis genes in never-smoker women with pulmonary adenocarcinoma. Cancer. 118:5588–5594. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Nakamura H, Kawasaki N, Taguchi M and Kabasawa K: Survival impact of epidermal growth factor receptor overexpression in patients with non-small cell lung cancer: A meta-analysis. Thorax. 61:140–145. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Takeda M, Okamoto I, Hirabayashi N, Kitano M and Nakagawa K: Thymidylate synthase and dihydropyrimidine dehydrogenase expression levels are associated with response to S-1 plus carboplatin in advanced non-small cell lung cancer. Lung Cancer. 73:103–109. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Honma Y, Togo S, Shimizu K, Tulafu M, Hayashi T, Uekusa T, Tominaga S, Kido K, Fujimoto Y, Nanba Y, et al: Expression of thymidylate synthase predicts clinical outcomes of S-1-based chemotherapy in squamous cell lung cancer. Oncol Lett. 14:3319–3326. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Liu Q, Yu Z, Xiang Y, Wu N, Wu L, Xu B, Wang L, Yang P, Li Y and Bai L: Prognostic and predictive significance of thymidylate synthase protein expression in non-small cell lung cancer: A systematic review and meta-analysis. Cancer Biomark. 15:65–78. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Huang CL, Yokomise H, Kobayashi S, Fukushima M, Hitomi S and Wada H: Intratumoral expression of thymidylate synthase and dihydropyrimidine dehydrogenase in non-small cell lung cancer patients treated with 5-FU-based chemotherapy. Int J Oncol. 17:47–54. 2000.PubMed/NCBI | |
|
Nakano J, Huang C, Liu D, Masuya D, Nakashima T, Yokomise H, Ueno M, Wada H and Fukushima M: Evaluations of biomarkers associated with 5-FU sensitivity for non-small-cell lung cancer patients postoperatively treated with UFT. Br J Cancer. 95:607–615. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Lenz HJ, Leichman CG, Danenberg KD, Danenberg PV, Groshen S, Cohen H, Laine L, Crookes P, Silberman H, Baranda J, et al: Thymidylate synthase mRNA level in adenocarcinoma of the stomach: A predictor for primary tumor response and overall survival. J Clin Oncol. 14:176–182. 1996.PubMed/NCBI View Article : Google Scholar | |
|
Okabe T, Okamoto I, Tsukioka S, Uchida J, Hatashita E, Yamada Y, Yoshida T, Nishio K, Fukuoka M, Jänne PA and Nakagawa K: Addition of S-1 to the epidermal growth factor receptor inhibitor gefitinib overcomes gefitinib resistance in non-small cell lung cancer cell lines with MET amplification. Clin Cancer Res. 15:907–913. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Okabe T, Okamoto I, Tsukioka S, Uchida J, Iwasa T, Yoshida T, Hatashita E, Yamada Y, Satoh T, Tamura K, et al: Synergistic antitumor effect of S-1 and the epidermal growth factor receptor inhibitor gefitinib in non-small cell lung cancer cell lines: Role of gefitinib-induced down-regulation of thymidylate synthase. Mol Cancer Ther. 7:599–606. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Takezawa K, Okamoto I, Tanizaki J, Kuwata K, Yamaguchi H, Fukuoka M, Nishio K and Nakagawa K: Enhanced anticancer effect of the combination of BIBW2992 and thymidylate synthase-targeted agents in non-small cell lung cancer with the T790M mutation of epidermal growth factor receptor. Mol Cancer Ther. 9:1647–1656. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Sheng M, Wang F, Zhao Y, Li S, Wang X, Shou T, Luo Y and Tang W: Comparison of clinical outcomes of patients with non-small-cell lung cancer harbouring epidermal growth factor receptor exon 19 or exon 21 mutations after tyrosine kinase inhibitors treatment: A meta-analysis. Eur J Clin Pharmacol. 72:1–11. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Eck MJ and Yun CH: Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta. 1804:559–566. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N and Sliwkowski MX: Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 66:8163–8171. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Okabe T, Okamoto I, Tamura K, Terashima M, Yoshida T, Satoh T, Takada M, Fukuoka M and Nakagawa K: Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res. 67:2046–2053. 2007.PubMed/NCBI View Article : Google Scholar | |
|
Cho J, Chen L, Sangji N, Okabe T, Yonesaka K, Francis JM, Flavin RJ, Johnson W, Kwon J, Yu S, et al: Cetuximab response of lung cancer-derived EGF receptor mutants is associated with asymmetric dimerization. Cancer Res. 73:6770–6779. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Imai H, Minemura H, Kishikawa T, Yamada Y, Suzuki K, Umeda Y, Wasamoto S, Kasahara N, Ishihara S, Yamaguchi O, et al: Efficacy and safety of S-1 monotherapy in previously treated elderly patients (aged ≥75 years) with non-small cell lung cancer: A retrospective analysis. Thorac Cancer. 11:2867–2876. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Tsuchiya T, Arai J, Matsumoto K, Miyazaki T, Honda S, Tagawa T, Nakamura A, Taniguchi H, Sano I, Akamine S, et al: Prognostic impact of the ABCC11/MRP8 polymorphism in adjuvant oral chemotherapy with S-1 for non-small cell lung cancer. Chemotherapy. 61:77–86. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Uemura T, Oguri T, Maeno K, Sone K, Takeuchi A, Fukuda S, Kunii E, Takakuwa O, Kanemitsu Y, Ohkubo H, et al: ABCC11 gene polymorphism as a potential predictive biomarker for an oral 5-fluorouracil derivative drug S-1 treatment in non-small cell lung cancer. Cancer Chemother Pharmacol. 84:1229–1239. 2019.PubMed/NCBI View Article : Google Scholar |