Mendelian randomization analysis provides insights into the relationship between inflammatory bowel disease and skin cancer
- Authors:
- Published online on: July 10, 2025 https://doi.org/10.3892/mco.2025.2878
- Article Number: 83
-
Copyright: © Rashid et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
The incidence of numerous multifactorial diseases, including skin cancer and inflammatory bowel diseases, has steadily increased worldwide over the past several decades (1-3). Previous studies have proposed an increased risk of both melanoma and non-melanoma skin cancers (NMSC) in patients with inflammatory bowel disease (IBD) (4-6). This association was previously considered to be a direct consequence of thiopurine therapy due to the increased generation of reactive oxygen species from 6-thioguanine (4,7). However, Lo et al (8) reported in their meta-analysis that patients with IBD demonstrate an increased risk of NMSC even when controlled for immunosuppressant use [Crohn's disease (CD) incidence rate ratio (IRR)=2.22; 95% confidence interval (CI): 1.41-3.48]; ulcerative colitis (UC) IRR=1.38; 95% CI: 1.12-1.71]. Genome-wide association studies (GWAS) have also identified numerous susceptibility loci associated with IBD, with some loci also implicated in cancer, but not skin cancer specifically (9). Although a clear biological explanation linking IBD with skin cancer has yet to be established, sustained cutaneous inflammation (10), immune suppression (11), and shared germline susceptibility are suspected to play a role (12).
While ultraviolet radiation (UVR)-induced mutagenesis remains to be the most well-established risk factor for skin cancer (13), emerging evidence suggests that neuro-immuno-endocrine mechanisms within the skin may contribute to skin cancer susceptibility (14-17). The neuro-immuno-endocrine functions of the skin involves a complex set of components and interactions spanning different organs, including the brain, gut and adrenal glands (14). Circulating immune cells, such as macrophages, lymphocytes and Langerhans cells, interact with cutaneous nerve fibers to modulate host defense, inflammation and tissue repair (18). Neuropeptides released from nerve terminals may further modulate the function of cutaneous immune cells, while immune-derived mediators can influence neuronal activity, contributing to sensations such as itch and pain (19). UVR-induced modulation of cytokines such as TNFα, IL-1 and IL-6 drive pathways such as the central HPA axis to generate immunosuppressive effects (20).
Vitamin D signaling, particularly through the vitamin D receptor (VDR), plays a significant role in melanoma progression and management (21,22). The VDR is a nuclear receptor activated by 1,25-dihydroxyvitamin D3 (calcitriol), which influences various cellular processes including proliferation, differentiation and immune responses. High VDR expression in melanoma cells is associated with improved outcomes and reduced melanoma-related mortality (22). This is partly due to the inhibition of the Wnt/β-catenin signaling pathway, which is known to promote melanoma progression (23). Despite increasing evidence unraveling the neuro-immuno-endocrine potential of the skin, current mechanisms remain to be poorly understood and are primarily derived from animal models and limited clinical studies.
Mendelian randomization (MR) is a powerful tool used to explore causal relationships between exposure traits and disease outcomes by utilizing genetic variants as proxies for the exposure of interest, thereby minimizing confounding and reverse causation (24). Over the past year, recent MR efforts for IBD and skin cancer have yielded mixed results, with some evidence to support causal effects for UC on NMSC in East Asian and European cohorts (25,26).
Given the growing availability of GWAS data and the need to clarify these associations, it was sought to determine whether there is a causal genetic relationship between IBD subtypes (CD and UC) and skin cancer (both melanoma and non-melanoma) using publicly available GWAS repositories. The present study aims to provide further insights into the potential shared genetic architecture between these conditions and to elucidate whether the observed associations are likely to be causal or a result of confounding factors.
Materials and methods
Instrument selection
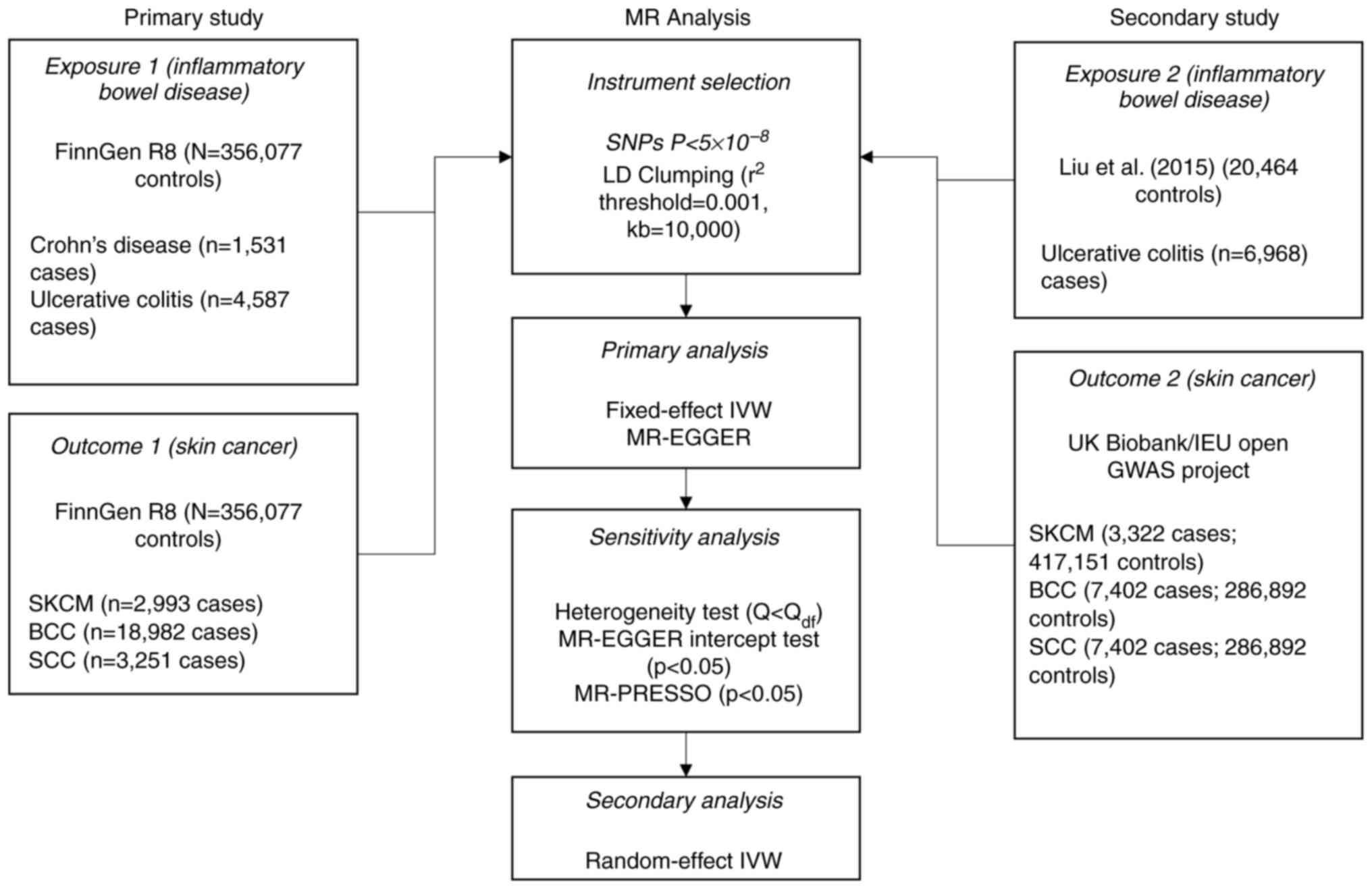
Summary statistics were gathered and imputed against a Finnish-specific whole genome sequence backbone containing 20,175,454 variants. From 356,077 total controls, summary statistics were retrieved from FinnGen R8 for: CD (1,531 cases), UC (4,857 cases), SKCM (2,993 cases), BCC (18,982 cases) and SCC (251 cases). Lead significant single nucleotide polymorphisms (SNPs) with genome-wide significance level (P<5x10-8) were then selected and linkage disequilibrium pruning (r2 threshold=0.001, clumping window=10 Kb) was employed to distinguish highly correlated SNPs. Ethics approval was not required for the present study because no individual-level data were used in summary statistics.
To assess the robustness of the present findings, skin cancer GWAS summary statistics for significant MR associations were also retrieved from the UK Biobank (UKBB) consortium representing self-reported and code-verified cases of SKCM (3,322 cases; 417,151 controls), BCC (7,402 cases; 286,892 controls) and SCC (7,402 cases; 286,892 controls) as part of a secondary study. Summary statistics for UC in the secondary study were retrieved from a 2015 meta-analysis of European ancestry containing 6,968 cases and 20,464 controls (9).
Data preparation
Data preparation and MR analysis was performed using the TwoSampleMR (27) package (Version 0.5.6) within R. Instrumental variables were harmonized to matching effect alleles with palindromic SNPs excluded. The fixed-effect inverse variance weighted (IVW) method was selected to approximate exposure effects on outcome. The IVW method assumes all genetic variants are instrumental variables, and is a common estimator used in two-sample MR (28). MR causal effects (β) were considered significant at a Bonferroni-corrected P-value 0.05/3=0.017, with P-values >0.017, with P<0.05 considered as a suggestive association.
Sensitivity analyses
Sensitivity analysis were used in the present study to assess whether our results violated key MR assumptions (29). Importantly, genetic instruments should not be associated with potential confounding variables. The MR-EGGER intercept test was used to assess whether the fixed-effect IVW estimate is biased by the presence of directional pleiotropy. To further check for the presence of directional pleiotropy, MR pleiotropy residual sum and outlier (MR-PRESSO) estimates were subsequently incorporated into our sensitivity analysis. For this test, the global test P-value detects for the presence of horizontal pleiotropy in the IVW estimate. If horizontal pleiotropy was detected in the first step, then MR-PRESSO corrected the raw IVW estimate to mitigate outlier effects. Heterogeneity calculations were also performed to assess the stability and consistency of SNP effects across analyses.
Results
Genetic associations for IBD and three major types of skin cancer [cutaneous melanoma (SKCM), basal cell carcinoma (BCC) and squamous cell carcinoma (SCC)] were retrieved from FinnGen Release 8 (R8) and subjected to two-sample MR. There were a total of 371 significantly-associated variants for CD and 3,193 variants for UC available for MR. An overview for genetic instrument selection is shown in Fig. 1.
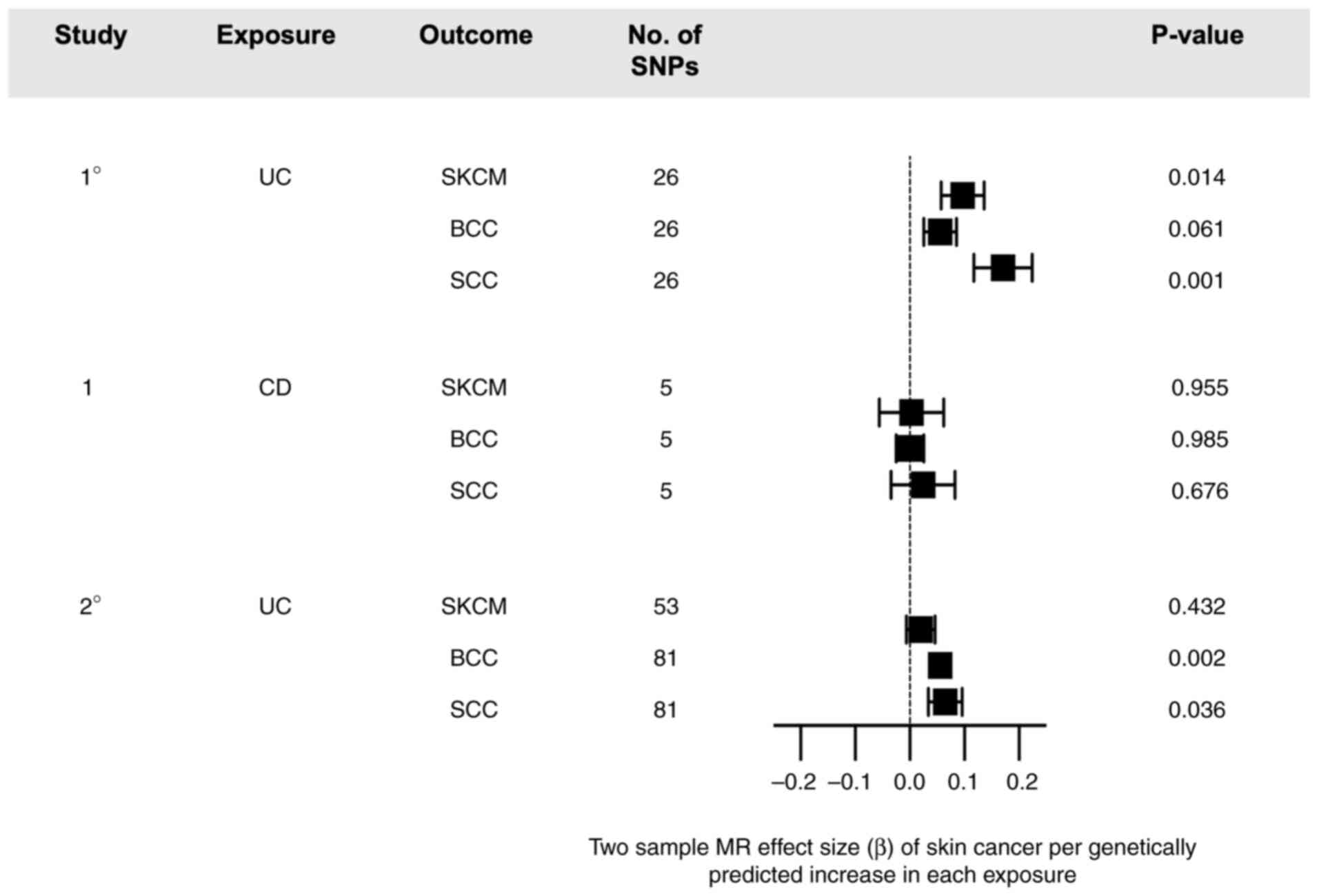
There were 26 significant SNPs associated with UC as an exposure post-harmonization (Fig. 2). Fixed-effect IVW regression showed significant causal effects for SKCM (beta=0.097, se=0.039, P=0.0138) and SCC (beta=0.171, se=0.054, P=0.0014), but not BCC (beta=0.056, se=0.030, P=0.0610) (Table SI). A causal association was also observed using MR-EGGER regression, although MR estimates were found to lack significance. Using Cochran's Q test, the presence of heterogeneity was observed for SKCM (Q=27.043, P=0.302), which was shown to increase for SCC (Q=51.914, P=8.03x10-4) (Table I). Due to presence of heterogeneity, a random-effect IVW analysis was performed and showed a consistent association for SKCM (beta=0.0862, P=0.0157) and SCC (OR=0.174, P=6.57x10-4). For SKCM, the MR-EGGER intercept test showed presence of directional pleiotropy (intercept=-0.0058, P<2.2x10-16) with no outlier correction using MR-PRESSO. For SCC, there was also presence of directional pleiotropy (intercept=-0.068, P<2.2x10-16) with outlier correction (OR=0.0392, P=0.102) showing no significant association.
Table IResults of heterogeneity testing and sensitivity analyses for primary study genetic instruments. |
Using CD as an exposure, only 5 significant SNPs were included post-harmonization for MR analysis. MR using IVW regression demonstrated no significant causal effects for SKCM (beta=0.003, se=0.059, P=0.955), BCC (beta=4.82x10-4, se=2.54x10-2, P=0.985), or SCC (beta=0.024, se=0.058, P=0.676). No heterogeneity was found using Cochran's Q test. Directional pleiotropy was again detected for SKCM (intercept=0.004, P=8.93x10-3) and SCC (intercept=-0.054, P=3.62x10-5), with no outlier correction using MR-PRESSO.
To test the reproducibility of causal effects observed for UC, GWAS summary statistics from Liu et al (9) and the UK Biobank (Fig. 1) were utilized for a secondary study. A total of 83 significant SNPs were found post-harmonization for SKCM, and 81 significant SNPs found for both BCC and SCC. MR analysis demonstrated suggestive or significant causal effects between UC and SCC (beta=0.065, se=0.031, P=0.036) and UC and BCC (beta=0.056, se=0.018, P=0.002), but not UC and SKCM (beta=0.020, se=0.026, P=0.432).
Discussion
The current analysis indicates causal effects for UC on SCC, which is supported by a recent meta-GWAS East Asian and European ancestries (25). Secondary analysis using datasets from the UK Biobank replicated the significant associations for UC and SCC, providing additional evidence for this relationship using European cohorts. Utilization of the FinnGen R8 dataset in the present study offered a high prevalence of cases relative to total population compared with other publicly available datasets. Moreover, imputation with Finnish-specific panels has also been shown to demonstrate high accuracy for low-frequency variants (30). The influence of confounding bias was controlled for using strict selection of instrumental variables and a priori sensitivity analyses. Sensitivity testing showed moderate heterogeneity with possible pleiotropy for the UC and skin cancer associations. Ultimately, these findings suggest that select genetic variants may influence skin cancer through pathways unrelated to UC, although outlier correction was applied to account for the observed pleiotropy.
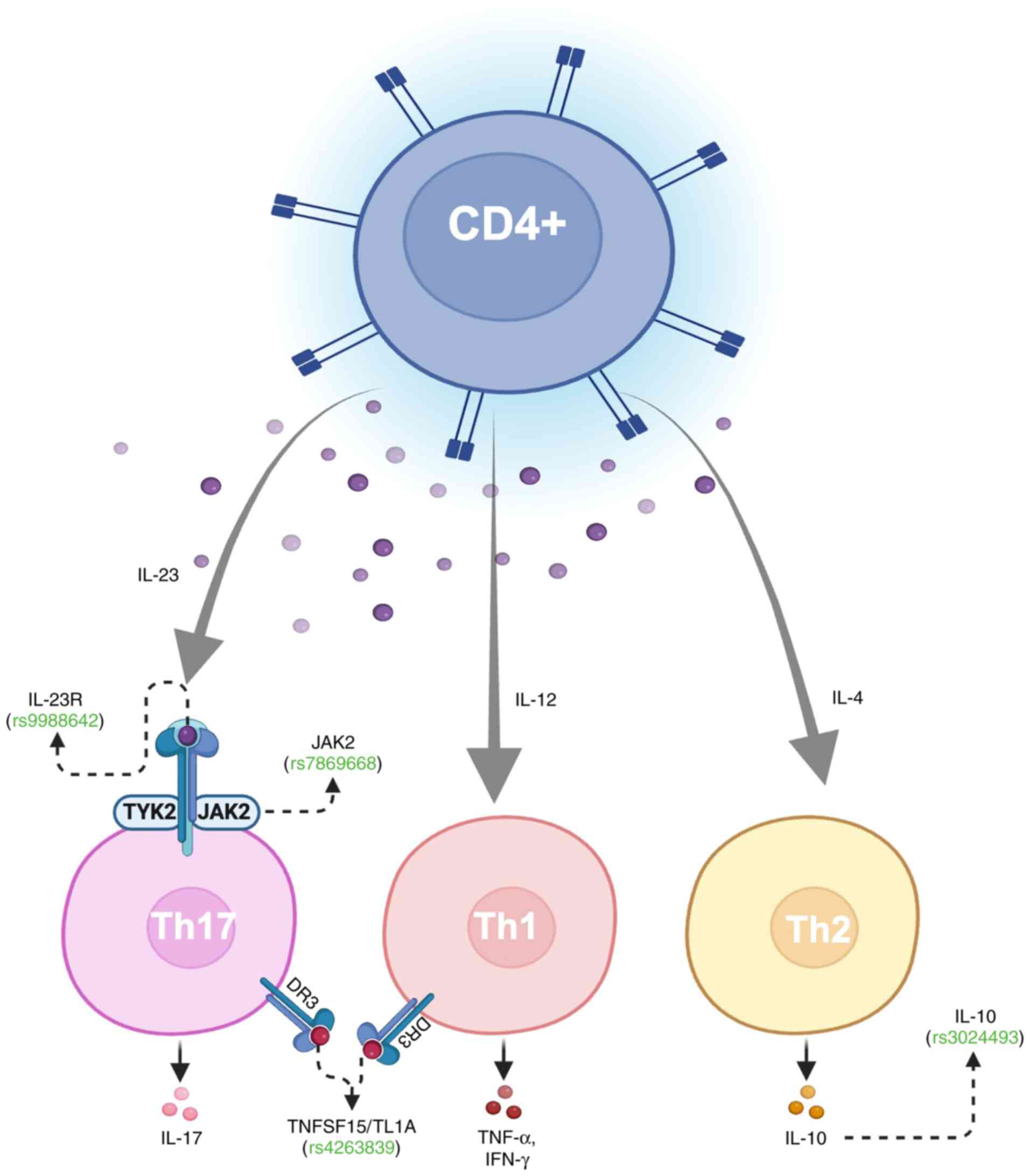
There are a few noteworthy observations from the 26 UC-associated SNPs used for MR (Table SII). First, rs3024493, proximal to interleukin 10 (IL-10), has been linked to the development of IBD in genetic studies (31,32). IL-10 is an important immunoregulator of mucosal immunity which acts by inhibiting proinflammatory cytokines (IL-1β, TNF-α and IL-6), Th2 cell-derived cytokines (IL-4 and IL-5), and chemokines (MIP-1α and interleukin-8), while increasing synthesis of several anti-inflammatory proteins (33). In patients with melanoma, rs3024493 has been linked with downregulation of IL-10 secretion in CD4+ T cells. Multivariate analyses have demonstrated decreased melanoma overall survival [hazard ratio (HR): 4.73; 95% CI: 1.68-13.29] for minor allele homozygotes (TT) compared with major allele homozygotes (GG), although interestingly show improved overall survival (HR: 0.58, 95% CI: 0.39-0.86) in heterozygotes (GT). Conversely, increased production of IL-10 in keratinocyte carcinomas has been hypothesized as a mechanism for evading local T cell-mediated immune responses (34). This may suggest that the regulatory function of IL-10 extends to skin immunity and cancer progression. Given that UV radiation modulates cytokine expression, including IL-10, it is plausible that chronic UV exposure may may provide a potential link between chronic inflammation and immune escape mechanisms in skin cancer (35).
More broadly, the IL-23/Th-17 axis for inflammation has been implicated in both IBD (36) and skin cancers (37,38). When bound to IL-23R (rs9988642), IL-23 activates JAK2 (rs7869668) to facilitate differentiation of naïve CD4+ T-cells into IL-17-secreting Th17 cells (Fig. 3). This differentiation process establishes a pro-inflammatory microenvironment which favors tumorigenesis. TL1A (rs4263839) interacts with DR3 to synergistically increase the production of IL-17 and other pro-inflammatory cytokine as observed in CD (39,40). IL-17 then acts to promote infiltration of myeloid-derived suppressor cells and activate the STAT3 signaling pathway, which has been canonically associated with increased tumor cell proliferation, survival, and angiogenesis (41-43). Together, these interactions collectively reinforce the IL-23/Th17 axis as a potential driver of inflammation-mediated tumorigenesis.
The skin and gut function as the two largest immune systems in the human body, each employing distinct yet overlapping biological mechanisms (44). Dysbiosis in the gut microbiota drives biological crosstalk in what is known as the skin-gut axis, in which cell trafficking between the skin and gut promotes frequent comorbidity of inflammatory diseases (45). Recent findings suggest that gut-derived metabolites (for example, short-chain fatty acids, bile acids and vitamins) influence skin barrier function (46-48), while skin-derived inflammatory mediators may exacerbate gut inflammation (49,50). Furthermore, immune cell trafficking between the gut and skin is increasingly recognized as a mechanism for disease spread (45). Aberrant migration of gut-educated T cells has been implicated in both cutaneous and gastrointestinal inflammation (51,52), even extending to other inflammatory diseases such as ankylosing spondylitis and primary sclerosing cholangitis (52,53).
While gut and skin leukocytes are typically directed to their respective tissues, the presence of inflammation may alter homing patterns. For instance, intestinal T cells may acquire skin-homing characteristics during chronic gut inflammation, leading to secondary cutaneous disease (54). Conversely, skin-resident memory T cells may enter circulation and contribute to systemic inflammation, as observed in graft-vs.-host disease (55). These insights also have therapeutic implications for modulating gut-skin immune cell trafficking. For example, the use of vedolizumab, a gut-selective integrin blocker used in IBD, has been hypothesized to modulate binding of α4β7-MAdCAM1 for gut access and subsequent skin entry (56).
The present study has several important limitations. First, the limited number of CD cases (1,531 in FinnGen R8) resulted in the identification of only five genome-wide significant SNPs available for MR analysis post-harmonization. This small number of instruments significantly weakened the strength of the genetic proxies for CD, therefore increasing the likelihood of weak instrument bias. As a result, no significant causal associations were observed between CD and skin cancer outcomes in our MR analyses. Additionally, while the use of Finnish-specific GWAS data enhances variant detection, the findings may not be fully generalizable to other populations with more diverse genetic backgrounds. Finally, the presence of pleiotropy, as suggested by the MR-EGGER intercept tests, indicates that some genetic variants might influence skin cancer risk through mechanisms that are independent of IBD.
Overall, the present study demonstrates a causal genetic association between UC and SCC in European cohorts distinct from biologic-induced skin cancer in previous literature. The demonstrated genetic associations may provide meaningful implications for clinical practice, particularly in terms of risk stratification, preventative counseling and surveillance strategies for patients with IBD. While UV exposure remains the predominant risk factor for SCC, IBD-related immune dysregulation may create a permissive environment for carcinogenesis. Patients with UC, particularly those with longstanding disease, may benefit from personalized preventative strategies, including education on sun safety, regular dermatologic screenings, and early intervention measures. Further investigation of the IL-23/Th17 axis in UC-associated SCC may elucidate underlying disease mechanisms and inform the development of new targeted therapeutic strategies.
Supplementary Material
Calculated MR effects of IBD on skin cancers using five different MR methods: MR Egger, Weighted median, Inverse variance weighted, Simple mode and Weighted mode.
Final list of instrumental variables post-harmonization for UC used for mendelian randomization analysis. Annotations are based on the latest build of the human genome (GRCh38.p14) using the Single Nucleotide Polymorphism Database (dbSNP) and may be subject to updates and revisions. rsID, Reference SNP cluster ID.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be found at the following URLs: FinnGen (https://www.finngen.fi/en), UK Biobank (https://www.ukbiobank.ac.uk/), and the IEU Open GWAS Project (https://gwas.mrcieu.ac.uk/).
Authors' contributions
SR, DU, SG, MA and HT conceptualized and designed the study, conducted formal analysis, prepared the original draft, and wrote, reviewed and edited the manuscript. SR, MA and HT curated data. HT acquired funding. All authors read and approved the final version of the manuscript. SR and HT confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Leiter U, Keim U and Garbe C: Epidemiology of skin cancer: Update 2019. Adv Exp Med Biol. 1268:123–139. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Burisch J and Munkholm P: Inflammatory bowel disease epidemiology. Curr Opin Gastroenterol. 29:357–362. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al: Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet. 390:2769–2778. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Singh S, Nagpal SJS, Murad MH, Yadav S, Kane SV, Pardi DS, Talwalkar JA and Loftus EV Jr: Inflammatory bowel disease is associated with an increased risk of melanoma: A systematic review and meta-analysis. Clin Gastroenterol Hepatol. 12:210–218. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Narous M, Nugent Z, Singh H and Bernstein CN: Risks of melanoma and nonmelanoma skin cancers pre- and post-inflammatory bowel disease diagnosis. Inflamm Bowel Dis. 29:1047–1056. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Long MD, Herfarth HH, Pipkin CA, Porter CQ, Sandler RS and Kappelman MD: Increased risk for non-melanoma skin cancer in patients with inflammatory bowel disease. Clin Gastroenterol Hepatol. 8:268–274. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Brem R, Li F and Karran P: Reactive oxygen species generated by thiopurine/UVA cause irreparable transcription-blocking DNA lesions. Nucleic Acids Res. 37:1951–1961. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Lo B, Zhao M, Vind I and Burisch J: The risk of extraintestinal cancer in inflammatory bowel disease: A systematic review and meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 19:1117–1138.e19. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al: Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 47:979–986. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Marzano AV, Borghi A, Stadnicki A, Crosti C and Cugno M: Cutaneous manifestations in patients with inflammatory bowel diseases: Pathophysiology, clinical features, and therapy. Inflamm Bowel Dis. 20:213–227. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Long MD, Martin CF, Pipkin CA, Herfarth HH, Sandler RS and Kappelman MD: Risk of melanoma and nonmelanoma skin cancer among patients with inflammatory bowel disease. Gastroenterology. 143:390–399.e1. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Cushing KC, Du X, Chen Y, Stetson LC, Kuppa A, Chen VL, Kahlenberg JM, Gudjonsson JE, Vanderwerff B, Higgins PDR and Speliotes EK: Inflammatory bowel disease risk variants are associated with an increased risk of skin cancer. Inflamm Bowel Dis. 28:1667–1676. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Narayanan DL, Saladi RN and Fox JL: Ultraviolet radiation and skin cancer. Int J Dermatol. 49:978–986. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Slominski AT, Slominski RM, Raman C, Chen JY, Athar M and Elmets C: Neuroendocrine signaling in the skin with a special focus on the epidermal neuropeptides. Am J Physiol Cell Physiol. 323:C1757–C1776. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Rangwala S and Tsai KY: Roles of the immune system in skin cancer. Br J Dermatol. 165:953–965. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Kalaora S, Nagler A, Wargo JA and Samuels Y: Mechanisms of immune activation and regulation: Lessons from melanoma. Nat Rev Cancer. 22:195–207. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Tobin DJ: Biochemistry of human skin-our brain on the outside. Chem Soc Rev. 35:52–67. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Tamari M, Ver Heul AM and Kim BS: Immunosensation: Neuroimmune cross talk in the skin. Annu Rev Immunol. 39:369–393. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Liu AW, Gillis JE, Sumpter TL and Kaplan DH: Neuroimmune interactions in atopic and allergic contact dermatitis. J Allergy Clin Immunol. 151:1169–1177. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Slominski RM, Chen JY, Raman C and Slominski AT: Photo-neuro-immuno-endocrinology: How the ultraviolet radiation regulates the body, brain, and immune system. Proc Natl Acad Sci USA. 121(e2308374121)2024.PubMed/NCBI View Article : Google Scholar | |
|
Slominski RM, Kim TK, Janjetovic Z, Brożyna AA, Podgorska E, Dixon KM, Mason RS, Tuckey RC, Sharma R, Crossman DK, et al: Malignant melanoma: An overview, new perspectives, and vitamin D signaling. Cancers (Basel). 16(2262)2024.PubMed/NCBI View Article : Google Scholar | |
|
Muralidhar S, Filia A, Nsengimana J, Poźniak J, O'Shea SJ, Diaz JM, Harland M, Randerson-Moor JA, Reichrath J, Laye JP, et al: Vitamin D-VDR signaling inhibits Wnt/β-catenin-mediated melanoma progression and promotes antitumor immunity. Cancer Res. 79:5986–5998. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Kovacs D, Migliano E, Muscardin L, Silipo V, Catricalà C, Picardo M and Bellei B: The role of Wnt/β-catenin signaling pathway in melanoma epithelial-to-mesenchymal-like switching: Evidences from patients-derived cell lines. Oncotarget. 7:43295–43314. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Smith GD and Ebrahim S: ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 32:1–22. 2003.PubMed/NCBI View Article : Google Scholar | |
|
Liu D, Cao M, Wang H, Cao W, Zheng C, Li Y and Wang Y: Association between inflammatory bowel disease and cancer risk: Evidence triangulation from genetic correlation, Mendelian randomization, and colocalization analyses across East Asian and European populations. BMC Med. 22(137)2024.PubMed/NCBI View Article : Google Scholar | |
|
Li A, Yu M, Wu K, Liu L and Sun X: Inflammatory bowel disease and skin cancer: A two-sample mendelian randomization analysis. Genet Test Mol Biomarkers. 28:91–99. 2024.PubMed/NCBI View Article : Google Scholar | |
|
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al: The MR-Base platform supports systematic causal inference across the human phenome. Elife. 7(e34408)2018.PubMed/NCBI View Article : Google Scholar | |
|
Burgess S, Foley CN, Allara E, Staley JR and Howson JMM: A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 11(376)2020.PubMed/NCBI View Article : Google Scholar | |
|
Greenland S: An introduction to instrumental variables for epidemiologists. Int J Epidemiol. 29:722–729. 2000.PubMed/NCBI View Article : Google Scholar | |
|
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, et al: FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 613:508–518. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D, et al: Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 40:1319–1323. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Wang AH, Lam WJ, Han DY, Ding Y, Hu R, Fraser AG, Ferguson LR and Morgan AR: The effect of IL-10 genetic variation and interleukin 10 serum levels on Crohn's disease susceptibility in a New Zealand population. Hum Immunol. 72:431–435. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Li MC and He SH: IL-10 and its related cytokines for treatment of inflammatory bowel disease. World J Gastroenterol. 10:620–625. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Kim J, Modlin RL, Moy RL, Dubinett SM, McHugh T, Nickoloff BJ and Uyemura K: IL-10 production in cutaneous basal and squamous cell carcinomas. A mechanism for evading the local T cell immune response. J Immunol. 155:2240–2247. 1995.PubMed/NCBI | |
|
Enk CD, Sredni D, Blauvelt A and Katz SI: Induction of IL-10 gene expression in human keratinocytes by UVB exposure in vivo and in vitro. J Immunol. 154:4851–4856. 1995.PubMed/NCBI | |
|
Cătană CS, Berindan Neagoe I, Cozma V, Magdaş C, Tăbăran F and Dumitraşcu DL: Contribution of the IL-17/IL-23 axis to the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 21:5823–5830. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Ganzetti G, Rubini C, Campanati A, Zizzi A, Molinelli E, Rosa L, Simonacci F and Offidani A: IL-17, IL-23, and p73 expression in cutaneous melanoma: A pilot study. Melanoma Res. 25:232–238. 2015.PubMed/NCBI View Article : Google Scholar | |
|
McAllister F and Kolls JK: Th17 cytokines in non-melanoma skin cancer. Eur J Immunol. 45:692–694. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Kamada N, Hisamatsu T, Honda H, Kobayashi T, Chinen H, Takayama T, Kitazume MT, Okamoto S, Koganei K, Sugita A, et al: TL1A produced by lamina propria macrophages induces Th1 and Th17 immune responses in cooperation with IL-23 in patients with Crohn's disease. Inflamm Bowel Dis. 16:568–575. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Takedatsu H, Michelsen KS, Wei B, Landers CJ, Thomas LS, Dhall D, Braun J and Targan SR: TL1A (TNFSF15) regulates the development of chronic colitis by modulating both T-helper 1 and T-helper 17 activation. Gastroenterology. 135:552–567. 2008.PubMed/NCBI View Article : Google Scholar | |
|
He D, Li H, Yusuf N, Elmets CA, Li J, Mountz J and Xu H: IL-17 promotes tumor development through the induction of tumor promoting microenvironments at tumor sites and myeloid-derived suppressor cells. J Immunol. 184:2281–2288. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Wang L, Yi T, Zhang W, Pardoll DM and Yu H: IL-17 enhances tumor development in carcinogen-induced skin cancer. Cancer Res. 70:10112–10120. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Zou S, Tong Q, Liu B, Huang W, Tian Y and Fu X: Targeting STAT3 in cancer immunotherapy. Mol Cancer. 19(145)2020.PubMed/NCBI View Article : Google Scholar | |
|
Zhang J and Yao Z: Immune cell trafficking: A novel perspective on the gut-skin axis. Inflamm Regen. 44(21)2024.PubMed/NCBI View Article : Google Scholar | |
|
Galván-Peña S, Zhu Y, Hanna BS, Mathis D and Benoist C: A dynamic atlas of immunocyte migration from the gut. Sci Immunol. 9(eadi0672)2024.PubMed/NCBI View Article : Google Scholar | |
|
Brauckmann V, Nambiar S, Potthoff A, Höxtermann S, Wach J, Kayser A, Tiemann C, Schuppe AK, Brockmeyer NH and Skaletz-Rorowski A: Influence of dietary supplementation of short-chain fatty acid sodium propionate in people living with HIV (PLHIV). J Eur Acad Dermatol Venereol. 36:881–889. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Shi Z, Wu X, Wu CY, Singh SP, Law T, Yamada D, Huynh M, Liakos W, Yang G, Farber JM, et al: Bile acids improve psoriasiform dermatitis through inhibition of IL-17A expression and CCL20-CCR6-mediated trafficking of T cells. J Invest Dermatol. 142:1381–1390.e11. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Trompette A, Pernot J, Perdijk O, Alqahtani RAA, Domingo JS, Camacho-Muñoz D, Wong NC, Kendall AC, Wiederkehr A, Nicod LP, et al: Gut-derived short-chain fatty acids modulate skin barrier integrity by promoting keratinocyte metabolism and differentiation. Mucosal Immunol. 15:908–926. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Dokoshi T, Seidman JS, Cavagnero KJ, Li F, Liggins MC, Taylor BC, Olvera J, Knight R, Chang JT, Salzman NH and Gallo RL: Skin inflammation activates intestinal stromal fibroblasts and promotes colitis. J Clin Invest. 131(e147614)2021.PubMed/NCBI View Article : Google Scholar | |
|
Leyva-Castillo JM, Galand C, Kam C, Burton O, Gurish M, Musser MA, Goldsmith JD, Hait E, Nurko S, Brombacher F, et al: Mechanical skin injury promotes food anaphylaxis by driving intestinal mast cell expansion. Immunity. 50:1262–1275.e4. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Adams DH and Eksteen B: Aberrant homing of mucosal T cells and extra-intestinal manifestations of inflammatory bowel disease. Nat Rev Immunol. 6:244–251. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Grant AJ, Lalor PF, Salmi M, Jalkanen S and Adams DH: Homing of mucosal lymphocytes to the liver in the pathogenesis of hepatic complications of inflammatory bowel disease. Lancet. 359:150–157. 2002.PubMed/NCBI View Article : Google Scholar | |
|
Salmi M and Jalkanen S: Human leukocyte subpopulations from inflamed gut bind to joint vasculature using distinct sets of adhesion molecules. J Immunol. 166:4650–4657. 2001.PubMed/NCBI View Article : Google Scholar | |
|
Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C and Song SY: Retinoic acid imprints gut-homing specificity on T cells. Immunity. 21:527–538. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Strobl J, Gail LM, Kleissl L, Pandey RV, Smejkal V, Huber J, Puxkandl V, Unterluggauer L, Dingelmaier-Hovorka R, Atzmüller D, et al: Human resident memory T cells exit the skin and mediate systemic Th2-driven inflammation. J Exp Med. 218(e20210417)2021.PubMed/NCBI View Article : Google Scholar | |
|
Colombel JF, Sands BE, Rutgeerts P, Sandborn W, Danese S, D'Haens G, Panaccione R, Loftus EV Jr, Sankoh S, Fox I, et al: The safety of vedolizumab for ulcerative colitis and Crohn's disease. Gut. 66:839–851. 2017.PubMed/NCBI View Article : Google Scholar |