The role of abnormal epigenetic regulation of small GTPases in glioma (Review)
- Authors:
- Mingyang Zhang
- Yimin Huang
- Qiang Zhang
- Xiaoyan Zhang
- Lumei Kang
- Jianguo Wang
-
Affiliations: Department of Histology and Embryology, Medical College, Nanchang University, Nanchang, Jiangxi 330006, P.R. China, Department of Central Laboratory, Jiaxing Women and Children's Hospital, Wenzhou Medical University, Jiaxing, Zhejiang 314000, P.R. China, Department of Clinical Laboratory, Longgang District People's Hospital of Shenzhen, Shenzhen, Guangdong 518172, P.R. China, Department of Medical Experimental Teaching Center, School of Basic Medical Sciences, Jiangxi Medical College, Nanchang University, Nanchang, Jiangxi 330006, P.R. China - Published online on: July 2, 2025 https://doi.org/10.3892/ijo.2025.5769
- Article Number: 63
-
Copyright: © Zhang et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
This article is mentioned in:
Abstract
 |
 |
 |
|
Price M, Neff C, Nagarajan N, Kruchko C, Waite KA, Cioffi G, Cordeiro BB, Willmarth N, Penas-Prado M, Gilbert MR, et al: CBTRUS statistical report: American brain tumor association & NCI system tumors neuro-oncology branch adolescent and young adult primary brain and other central nervous diagnosed in the United States in 2016-2020. Neuro Oncol. 26(Suppl 3): iii1–iii53. 2024. View Article : Google Scholar | |
|
Weller M, Wen PY, Chang SM, Dirven L, Lim M, Monje M and Reifenberger G: Glioma. Nat Rev Dis Primers. 10:332024. View Article : Google Scholar : PubMed/NCBI | |
|
Sanders S, Herpai DM, Rodriguez A, Huang Y, Chou J, Hsu FC, Seals D, Mott R, Miller LD and Debinski W: The presence and potential role of ALDH1A2 in the glioblastoma microenvironment. Cells. 10:24852021. View Article : Google Scholar : PubMed/NCBI | |
|
Schaff LR and Mellinghoff IK: Glioblastoma and other primary brain malignancies in adults: A Review. JAMA. 329:574–587. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Yasinjan F, Xing Y, Geng H, Guo R, Yang L, Liu Z and Wang H: Immunotherapy: A promising approach for glioma treatment. Front Immunol. 14:12556112023. View Article : Google Scholar : PubMed/NCBI | |
|
Xu S, Tang L, Li X, Fan F and Liu Z: Immunotherapy for glioma: Current management and future application. Cancer Lett. 476:1–12. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R and Reifenberger G: Glioma. Nat Rev Dis Primers. 1:150172015. View Article : Google Scholar : PubMed/NCBI | |
|
Wang LM, Englander ZK, Miller ML and Bruce JN: Malignant glioma. Adv Exp Med Biol. 1405:1–30. 2023. View Article : Google Scholar | |
|
Chen R, Smith-Cohn M, Cohen AL and Colman H: Glioma subclassifications and their clinical significance. Neurotherapeutics. 14:284–297. 2017. View Article : Google Scholar | |
|
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P and Ellison DW: The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, et al: The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 23:1231–1251. 2021. View Article : Google Scholar : | |
|
Luo Q, Liu Y, Yuan Z, Huang L and Diao B: Expression of Rab3b in human glioma: Influence on cell proliferation and apoptosis. Curr Pharm Des. 27:989–995. 2021. View Article : Google Scholar | |
|
Wang M, Jiang X, Yang Y, Chen H, Zhang C, Xu H, Qi B, Yao C and Xia H: Rhoj is a novel target for progression and invasion of glioblastoma by impairing cytoskeleton dynamics. Neurotherapeutics. 17:2028–2040. 2020. View Article : Google Scholar | |
|
Xu J, Galvanetto N, Nie J, Yang Y and Torre V: Rac1 promotes cell motility by controlling cell mechanics in human glioblastoma. Cancers (Basel). 12:16672020. View Article : Google Scholar | |
|
Reiner DJ and Lundquist EA: Small GTPases. WormBook. 2018:1–65. 2018. View Article : Google Scholar | |
|
Simanshu DK, Nissley DV and McCormick F: RAS proteins and their regulators in human disease. Cell. 170:17–33. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Ridley AJ: Life at the leading edge. Cell. 145:1012–1022. 2011. View Article : Google Scholar | |
|
Cherfils J and Zeghouf M: Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 93:269–309. 2013. View Article : Google Scholar | |
|
Chen K, Zhang Y, Qian L and Wang P: Emerging strategies to target RAS signaling in human cancer therapy. J Hematol Oncol. 14:1162021. View Article : Google Scholar : PubMed/NCBI | |
|
Zuo T, Wong S, Buza N and Hui P: KRAS mutation of extraovarian implants of serous borderline tumor: Prognostic indicator for adverse clinical outcome. Mod Pathol. 31:350–357. 2018. View Article : Google Scholar | |
|
Zhao S, Li Z, Zhang M, Zhang L, Zheng H, Ning J, Wang Y, Wang F, Zhang X, Gan H, et al: A brain somatic RHEB doublet mutation causes focal cortical dysplasia type II. Exp Mol Med. 51:1–11. 2019. | |
|
Gómez Del Pulgar T, Valdés-Mora F, Bandrés E, Pérez-Palacios R, Espina C, Cejas P, García-Cabezas MA, Nistal M, Casado E, González-Barón M, et al: Cdc42 is highly expressed in colorectal adenocarcinoma and downregulates ID4 through an epigenetic mechanism. Int J Oncol. 33:185–193. 2008. | |
|
Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M and Lengyel E: Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene. 19:3013–3020. 2000. View Article : Google Scholar : PubMed/NCBI | |
|
Li XR, Ji F, Ouyang J, Wu W, Qian LY and Yang KY: Overexpression of RhoA is associated with poor prognosis in hepatocellular carcinoma. Eur J Surg Oncol. 32:1130–1134. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Mazieres J, Antonia T, Daste G, Muro-Cacho C, Berchery D, Tillement V, Pradines A, Sebti S and Favre G: Loss of RhoB expression in human lung cancer progression. Clin Cancer Res. 10:2742–2750. 2004. View Article : Google Scholar | |
|
Shimada K, Uzawa K, Kato M, Endo Y, Shiiba M, Bukawa H, Yokoe H, Seki N and Tanzawa H: Aberrant expression of RAB1A in human tongue cancer. Br J Cancer. 92:1915–1921. 2005. View Article : Google Scholar | |
|
Li Y, Sun X, Ji D, Kong X, Liu D, Zhao Z, Yan J and Chen S: Expression of Rab5a correlates with tumor progression in pancreatic carcinoma. Virchows Arch. 470:527–536. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Hu X, Yin J, He R, Chao R and Zhu S: Circ_KCNQ5 participates in the progression of childhood acute myeloid leukemia by enhancing the expression of RAB10 via binding to miR-622. Hematology. 27:431–440. 2022. View Article : Google Scholar | |
|
Ge J, Chen Q, Liu B, Wang L, Zhang S and Ji B: Knockdown of Rab21 inhibits proliferation and induces apoptosis in human glioma cells. Cell Mol Biol Lett. 22:302017. View Article : Google Scholar : PubMed/NCBI | |
|
Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang JX, Huang XX, Cai MB, Tong ZT, Chen JW, Qian D, Liao YJ, Deng HX, Liao DZ, Huang MY, et al: Overexpression of the secretory small GTPase Rab27B in human breast cancer correlates closely with lymph node metastasis and predicts poor prognosis. J Transl Med. 10:2422012. View Article : Google Scholar : PubMed/NCBI | |
|
You X, Liu F, Zhang T, Li Y, Ye L and Zhang X: Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis. 34:1644–1652. 2013. View Article : Google Scholar | |
|
de Bruijn I, Kundra R, Mastrogiacomo B, Tran TN, Sikina L, Mazor T, Li X, Ochoa A, Zhao G, Lai B, et al: Analysis and visualization of longitudinal genomic and clinical data from the AACR Project GENIE biopharma collaborative in cBioPortal. Cancer Res. 83:3861–3867. 2023. View Article : Google Scholar : | |
|
Wang XF, Shi ZM, Wang XR, Cao L, Wang YY, Zhang JX, Yin Y, Luo H, Kang CS, Liu N, et al: MiR-181d acts as a tumor suppressor in glioma by targeting K-ras and Bcl-2. J Cancer Res Clin Oncol. 138:573–584. 2012. View Article : Google Scholar | |
|
Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W, Wang YY, Zhang JX, Jiang T, Kang CS, et al: Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro Oncol. 15:1491–1501. 2013. View Article : Google Scholar : | |
|
Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang C, Liu X, Wang X, Li H, Kang C, et al: MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol. 16:1341–1353. 2014. View Article : Google Scholar : | |
|
Zhang Y, Kim J, Mueller AC, Dey B, Yang Y, Lee DH, Hachmann J, Finderle S, Park DM, Christensen J, et al: Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B, and glioblastoma. Cell Death Differ. 21:720–734. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao Y, Pang D, Wang C, Zhong S and Wang S: MicroRNA-134 modulates glioma cell U251 proliferation and invasion by targeting KRAS and suppressing the ERK pathway. Tumour Biol. 37:11485–11493. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, Luo Z, Li G and Wu M: miR-181 subunits enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeleton remodeling in glioblastoma cells. Med Oncol. 31:8922014. View Article : Google Scholar : PubMed/NCBI | |
|
She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, Xiang J, Wu M and Li G: miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncol Rep. 32:957–964. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Li Z, Xu C, Ding B, Gao M, Wei X and Ji N: Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J Neurooncol. 134:19–28. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Wan J, Guo AA and Chowdhury I: TRPM7 induces mechanistic target of Rap1b through the downregulation of miR-28-5p in glioma proliferation and invasion. Front Oncol. 9:14132019. View Article : Google Scholar | |
|
Besse A, Sana J, Lakomy R, Kren L, Fadrus P, Smrcka M, Hermanova M, Jancalek R, Reguli S, Lipina R, et al: MiR-338-5p sensitizes glioblastoma cells to radiation through regulation of genes involved in DNA damage response. Tumour Biol. 37:7719–7727. 2016. View Article : Google Scholar | |
|
Kalhori MR, Irani S, Soleimani M, Arefian E and Kouhkan F: The effect of miR-579 on the PI3K/AKT pathway in human glioblastoma PTEN mutant cell lines. J Cell Biochem. 120:16760–16774. 2019. View Article : Google Scholar | |
|
Kalhori MR, Arefian E, Fallah Atanaki F, Kavousi K and Soleimani M: miR-548x and miR-4698 controlled cell proliferation by affecting the PI3K/AKT signaling pathway in Glioblastoma cell lines. Sci Rep. 10:15582020. View Article : Google Scholar : PubMed/NCBI | |
|
Schmidt N, Windmann S, Reifenberger G and Riemenschneider MJ: DNA hypermethylation and histone modifications downregulate the candidate tumor suppressor gene RRP22 on 22q12 in human gliomas. Brain Pathol. 22:17–25. 2012. View Article : Google Scholar | |
|
Shi C, Ren L, Sun C, Yu L, Bian X, Zhou X, Wen Y, Hua D, Zhao S, Luo W, et al: miR-29a/b/c function as invasion suppressors for gliomas by targeting CDC42 and predict the prognosis of patients. Br J Cancer. 117:1036–1047. 2017. View Article : Google Scholar | |
|
Sun G, Cao Y, Shi L, Sun L, Wang Y, Chen C, Wan Z, Fu L and You Y: Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother Radiopharm. 28:327–334. 2013.PubMed/NCBI | |
|
Qin W, Rong X, Dong J, Yu C and Yang J: miR-142 inhibits the migration and invasion of glioma by targeting Rac1. Oncol Rep. 38:1543–1550. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Tang H, Wang Z, Liu X, Liu Q, Xu G, Li G and Wu M: LRRC4 inhibits glioma cell growth and invasion through a miR-185-dependent pathway. Curr Cancer Drug Targets. 12:1032–1042. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Chen Q, Guo W, Zhang Y, Wu Y and Xiang J: MiR-19a promotes cell proliferation and invasion by targeting RhoB in human glioma cells. Neurosci Lett. 628:161–166. 2016. View Article : Google Scholar | |
|
Cai S, Shi CJ, Lu JX, Wang YP, Yuan T and Wang XP: miR-124-3p inhibits the viability and motility of glioblastoma multiforme by targeting RhoG. Int J Mol Med. 47:692021. View Article : Google Scholar | |
|
Quan Y, Song Q, Wang J, Zhao L, Lv J and Gong S: MiR-1202 functions as a tumor suppressor in glioma cells by targeting Rab1A. Tumour Biol. 39:10104283176975652017. View Article : Google Scholar : PubMed/NCBI | |
|
Xu D, Yu J, Gao G, Lu G, Zhang Y and Ma P: LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. 38:BSR201716642018. View Article : Google Scholar : | |
|
Fu Z, Luo W, Wang J, Peng T, Sun G, Shi J, Li Z and Zhang B: Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun. 492:480–486. 2017. View Article : Google Scholar | |
|
Gao W, Qiao M and Luo K: Long noncoding RNA TP53TG1 contributes to radioresistance of glioma cells via miR-524-5p/RAB5A axis. Cancer Biother Radiopharm. 36:600–612. 2021. | |
|
Zhang L, Chen H, Tian C and Zheng D: Propofol represses cell growth and metastasis by modulating the circular RNA Non-SMC condensin I complex subunit G/MicroRNA-200a-3p/RAB5A axis in glioma. World Neurosurg. 153:e46–e58. 2021. View Article : Google Scholar | |
|
Wu YJ, Yang QS, Chen H, Wang JT, Wang WB and Zhou L: Long non-coding RNA CASC19 promotes glioma progression by modulating the miR-454-3p/RAB5A axis and is associated with unfavorable MRI features. Oncol Rep. 45:728–737. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Shen G, Mao Y, Su Z, Du J, Yu Y and Xu F: PSMB8-AS1 activated by ELK1 promotes cell proliferation in glioma via regulating miR-574-5p/RAB10. Biomed Pharmacother. 122:1096582020. View Article : Google Scholar | |
|
Zhang X, Wang S, Lin G and Wang D: Down-regulation of circ-PTN suppresses cell proliferation, invasion and glycolysis in glioma by regulating miR-432-5p/RAB10 axis. Neurosci Lett. 735:1351532020. View Article : Google Scholar : PubMed/NCBI | |
|
Peng G, Su J, Xiao S and Liu Q: LINC00152 acts as a potential marker in gliomas and promotes tumor proliferation and invasion through the LINC00152/miR-107/RAB10 axis. J Neurooncol. 154:285–299. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Song D, Ye L, Xu Z, Jin Y and Zhang L: CircRNA hsa_ circ_0030018 regulates the development of glioma via regulating the miR-1297/RAB21 axis. Neoplasma. 68:391–403. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Xia Z, Liu F, Zhang J and Liu L: Decreased expression of MiRNA-204-5p contributes to glioma progression and promotes glioma cell growth, migration and invasion. PLoS One. 10:e01323992015. View Article : Google Scholar | |
|
Wang H, Wang Y, Bao Z, Zhang C, Liu Y, Cai J and Jiang C: Hypomethylated Rab27b is a progression-associated prognostic biomarker of glioma regulating MMP-9 to promote invasion. Oncol Rep. 34:1503–1509. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Q, Tang H, Liu X, Liao Y, Li H, Zhao Z, Yuan X and Jiang W: miR-200b as a prognostic factor targets multiple members of RAB family in glioma. Med Oncol. 31:8592014. View Article : Google Scholar | |
|
López-Ginés C, Navarro L, Muñoz-Hidalgo L, Buso E, Morales JM, Gil-Benso R, Gregori-Romero M, Megías J, Roldán P, Segura-Sabater R, et al: Association between epidermal growth factor receptor amplification and ADP-ribosylation factor 1 methylation in human glioblastoma. Cell Oncol (Dordr). 40:389–399. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
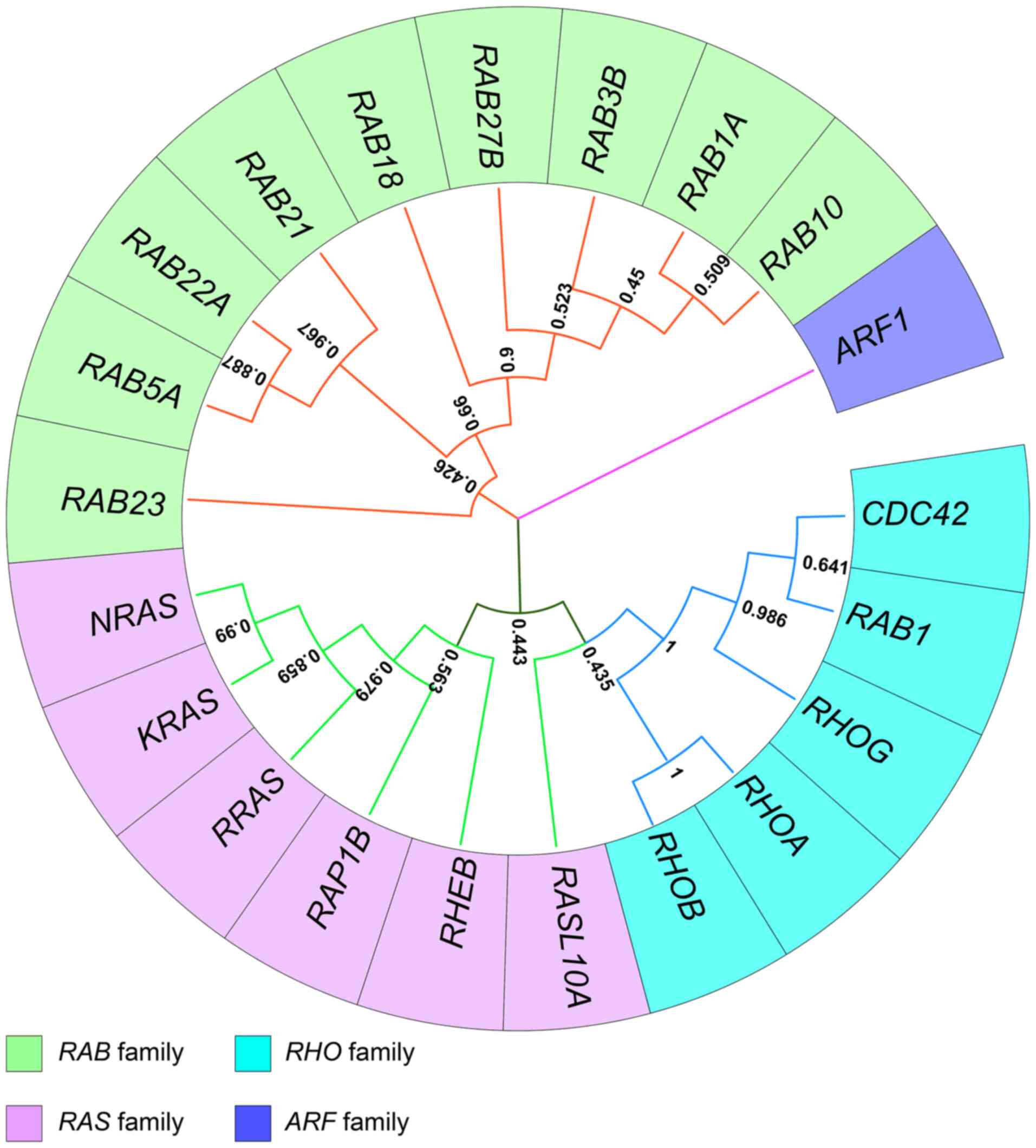
Tamura K, Stecher G and Kumar S: MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38:3022–3027. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Chen QW, Zhu XY, Li YY and Meng ZQ: Epigenetic regulation and cancer (review). Oncol Rep. 31:523–532. 2014. View Article : Google Scholar | |
|
Gu M, Ren B, Fang Y, Ren J, Liu X, Wang X, Zhou F, Xiao R, Luo X, You L and Zhao Y: Epigenetic regulation in cancer. MedComm (2020). 5:e4952024. View Article : Google Scholar : PubMed/NCBI | |
|
Zou Y and Xu H: Involvement of long noncoding RNAs in the pathogenesis of autoimmune diseases. J Transl Autoimmun. 3:1000442020. View Article : Google Scholar | |
|
Zong L, Zhou L, Hou Y, Zhang L, Jiang W, Zhang W, Wang L, Luo X, Wang S, Deng C, et al: Genetic and epigenetic regulation on the transcription of GABRB2: Genotype-dependent hydroxymethylation and methylation alterations in schizophrenia. J Psychiatr Res. 88:9–17. 2017. View Article : Google Scholar | |
|
Bai B, Chen S, Zhang Q, Jiang Q and Li H: Abnormal epigenetic regulation of the gene expression levels of Wnt2b and Wnt7b: Implications for neural tube defects. Mol Med Rep. 13:99–106. 2016. View Article : Google Scholar | |
|
Li P, Han M, Zhao X, Ren G, Mei S and Zhong C: Abnormal epigenetic regulations in the immunocytes of sjögren's syndrome patients and therapeutic potentials. Cells. 11:17672022. View Article : Google Scholar | |
|
Agirre X, Martínez-Climent JÁ, Odero M and Prosper F: Epigenetic regulation of miRNA genes in acute leukemia. Leukemia. 26:395–403. 2012. View Article : Google Scholar | |
|
Zeng C, Tsoi LC and Gudjonsson JE: Dysregulated epigenetic modifications in psoriasis. Exp Dermatol. 30:1156–1166. 2021. View Article : Google Scholar | |
|
Ramazi S, Dadzadi M, Sahafnejad Z and Allahverdi A: Epigenetic regulation in lung cancer. MedComm (2020). 4:e4012023. View Article : Google Scholar | |
|
Kondo Y, Katsushima K, Ohka F, Natsume A and Shinjo K: Epigenetic dysregulation in glioma. Cancer Sci. 105:363–369. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Lu Y, Chan YT, Tan HY, Li S, Wang N and Feng Y: Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol Cancer. 19:792020. View Article : Google Scholar : PubMed/NCBI | |
|
Robertson KD: DNA methylation and chromatin-unraveling the tangled web. Oncogene. 21:5361–5379. 2002. View Article : Google Scholar | |
|
Moore LD, Le T and Fan G: DNA methylation and its basic function. Neuropsychopharmacology. 38:23–38. 2013. View Article : Google Scholar | |
|
Illingworth RS and Bird AP: CpG islands-'a rough guide'. FEBS Lett. 583:1713–1720. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Antequera F: Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 60:1647–1658. 2003. View Article : Google Scholar | |
|
Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F and Esteller M: A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 66:8342–8346. 2006. View Article : Google Scholar | |
|
Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M and Cartron PF: Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenetics. 10:172018. View Article : Google Scholar : | |
|
Rasmussen KD and Helin K: Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30:733–750. 2016. View Article : Google Scholar : | |
|
Hatakeyama A, Hartmann B, Travers A, Nogues C and Buckle M: High-resolution biophysical analysis of the dynamics of nucleosome formation. Sci Rep. 6:273372016. View Article : Google Scholar | |
|
Torres IO and Fujimori DG: Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 35:68–75. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Wang G, Weng R, Lan Y, Guo X, Liu Q, Liu X, Lu C and Kang J: Synergetic effects of DNA methylation and histone modification during mouse induced pluripotent stem cell generation. Sci Rep. 7:395272017. View Article : Google Scholar : PubMed/NCBI | |
|
Ng MK and Cheung P: A brief histone in time: Understanding the combinatorial functions of histone PTMs in the nucleosome context. Biochem Cell Biol. 94:33–42. 2016. View Article : Google Scholar | |
|
Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ, Haukaas SA, Jonassen I, Petersen K, Goldfinger N, et al: Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 4:e46872009. View Article : Google Scholar : PubMed/NCBI | |
|
Tamura I, Maekawa R, Jozaki K, Ohkawa Y, Takagi H, Doi-Tanaka Y, Shirafuta Y, Mihara Y, Taketani T, Sato S, et al: Transcription factor C/EBPβ induces genome-wide H3K27ac and upregulates gene expression during decidualization of human endometrial stromal cells. Mol Cell Endocrinol. 520:1110852021. View Article : Google Scholar | |
|
Sungalee S, Liu Y, Lambuta RA, Katanayeva N, Donaldson Collier M, Tavernari D, Roulland S, Ciriello G and Oricchio E: Histone acetylation dynamics modulates chromatin conformation and allele-specific interactions at oncogenic loci. Nat Genet. 53:650–662. 2021. View Article : Google Scholar | |
|
Reda A, Hategan LA, McLean TAB, Creighton SD, Luo JQ, Chen SES, Hua S, Winston S, Reeves I, Padmanabhan A, et al: Role of the histone variant H2A.Z.1 in memory, transcription, and alternative splicing is mediated by lysine modification. Neuropsychopharmacology. 49:1285–1295. 2024. View Article : Google Scholar : PubMed/NCBI | |
|
Nemeth K, Bayraktar R, Ferracin M and Calin GA: Non-coding RNAs in disease: From mechanisms to therapeutics. Nat Rev Genet. 25:211–232. 2024. View Article : Google Scholar | |
|
Sana J, Faltejskova P, Svoboda M and Slaby O: Novel classes of non-coding RNAs and cancer. J Transl Med. 10:1032012. View Article : Google Scholar : | |
|
Brosnan CA and Voinnet O: The long and the short of noncoding RNAs. Curr Opin Cell Biol. 21:416–425. 2009. View Article : Google Scholar | |
|
Qin T, Li J and Zhang KQ: Structure, regulation, and function of linear and circular long non-coding RNAs. Front Genet. 11:1502020. View Article : Google Scholar : | |
|
Friedman RC, Farh KK, Burge CB and Bartel DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105. 2009. View Article : Google Scholar : | |
|
Shang R, Lee S, Senavirathne G and Lai EC: microRNAs in action: Biogenesis, function and regulation. Nat Rev Genet. 24:816–833. 2023. View Article : Google Scholar | |
|
Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R and Johnson R: Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. 19:535–548. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Khorkova O, Hsiao J and Wahlestedt C: Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 87:15–24. 2015. View Article : Google Scholar | |
|
Iwakiri J, Terai G and Hamada M: Computational prediction of lncRNA-mRNA interactionsby integrating tissue specificity in human transcriptome. Biol Direct. 12:152017. View Article : Google Scholar : PubMed/NCBI | |
|
Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M and Calin GA: Going circular: History, present, and future of circRNAs in cancer. Oncogene. 42:2783–2800. 2023. View Article : Google Scholar | |
|
Ma S, Kong S, Wang F and Ju S: CircRNAs: Biogenesis, functions, and role in drug-resistant Tumours. Mol Cancer. 19:1192020. View Article : Google Scholar : | |
|
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function as efficient microRNA sponges. Nature. 495:384–388. 2013. View Article : Google Scholar | |
|
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Yan H and Bu P: Non-coding RNA in cancer. Essays Biochem. 65:625–639. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Elam C, Hesson L, Vos MD, Eckfeld K, Ellis CA, Bell A, Krex D, Birrer MJ, Latif F and Clark GJ: RRP22 is a farnesylated, nucleolar, Ras-related protein with tumor suppressor potential. Cancer Res. 65:3117–3125. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Riemenschneider MJ, Reifenberger J and Reifenberger G: Frequent biallelic inactivation and transcriptional silencing of the DIRAS3 gene at 1p31 in oligodendroglial tumors with 1p loss. Int J Cancer. 122:2503–2510. 2008. View Article : Google Scholar | |
|
Tan Y, Zhang S, Xiao Q, Wang J, Zhao K, Liu W, Huang K, Tian W, Niu H, Lei T and Shu K: Prognostic significance of ARL9 and its methylation in low-grade glioma. Genomics. 112:4808–4816. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Rothhammer-Hampl T, Liesenberg F, Hansen N, Hoja S, Delic S, Reifenberger G and Riemenschneider MJ: Frequent epigenetic inactivation of DIRAS-1 and DIRAS-2 contributes to chemo-resistance in gliomas. Cancers (Basel). 13:51132021. View Article : Google Scholar | |
|
Bernal Astrain G, Nikolova M and Smith MJ: Functional diversity in the RAS subfamily of small GTPases. Biochem Soc Trans. 50:921–933. 2022. View Article : Google Scholar | |
|
Rásó E: Splice variants of RAS-translational significance. Cancer Metastasis Rev. 39:1039–1049. 2020. View Article : Google Scholar | |
|
Prior IA, Hood FE and Hartley JL: The frequency of ras mutations in cancer. Cancer Res. 80:2969–2974. 2020. View Article : Google Scholar | |
|
Knobbe CB, Reifenberger J and Reifenberger G: Mutation analysis of the Ras pathway genes NRAS, HRAS, KRAS and BRAF in glioblastomas. Acta Neuropathol. 108:467–470. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Wee P and Wang Z: Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel). 9:522017. View Article : Google Scholar : PubMed/NCBI | |
|
Sugiura R, Satoh R and Takasaki T: ERK: A double-edged sword in cancer. ERK-dependent apoptosis as a potential therapeutic strategy for cancer. Cells. 10:25092021. View Article : Google Scholar : PubMed/NCBI | |
|
Kim EK and Choi EJ: Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 1802:396–405. 2010. View Article : Google Scholar | |
|
An Z, Aksoy O, Zheng T, Fan QW and Weiss WA: Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene. 37:1561–1575. 2018. View Article : Google Scholar | |
|
Yang J, Yan J and Liu B: Targeting EGFRvIII for glioblastoma multiforme. Cancer Lett. 403:224–230. 2017. View Article : Google Scholar | |
|
Maruyama C, Tomisawa M, Wakana S, Yamazaki H, Kijima H, Suemizu H, Ohnishi Y, Urano K, Hioki K, Usui T, et al: Overexpression of human H-ras transgene is responsible for tumors induced by chemical carcinogens in mice. Oncol Rep. 8:233–237. 2001.PubMed/NCBI | |
|
Tsunematsu S, Saito H, Kagawa T, Morizane T, Hata J, Nakamura T, Ishii H, Tsuchiya M, Nomura T and Katsuki M: Hepatic tumors induced by carbon tetrachloride in transgenic mice carrying a human c-H-ras proto-oncogene without mutations. Int J Cancer. 59:554–559. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Weber SM and Carroll SL: The role of R-Ras proteins in normal and pathologic migration and morphologic change. Am J Pathol. 191:1499–1510. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Gutierrez-Erlandsson S, Herrero-Vidal P, Fernandez-Alfara M, Hernandez-Garcia S, Gonzalo-Flores S, Mudarra-Rubio A, Fresno M and Cubelos B: R-RAS2 overexpression in tumors of the human central nervous system. Mol Cancer. 12:1272013. View Article : Google Scholar : PubMed/NCBI | |
|
Nakada M, Niska JA, Tran NL, McDonough WS and Berens ME: EphB2/R-Ras signaling regulates glioma cell adhesion, growth, and invasion. Am J Pathol. 167:565–576. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang YL, Wang RC, Cheng K, Ring BZ and Su L: Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med. 14:90–99. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Altschuler D and Lapetina EG: Mutational analysis of the cAMP-dependent protein kinase-mediated phosphorylation site of Rap1b. J Biol Chem. 268:7527–7531. 1993. View Article : Google Scholar : PubMed/NCBI | |
|
Wittchen ES, Aghajanian A and Burridge K: Isoform-specific differences between Rap1A and Rap1B GTPases in the formation of endothelial cell junctions. Small GTPases. 2:65–76. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Lin KT, Yeh YM, Chuang CM, Yang SY, Chang JW, Sun SP, Wang YS, Chao KC and Wang LH: Glucocorticoids mediate induction of microRNA-708 to suppress ovarian cancer metastasis through targeting Rap1B. Nat Commun. 6:59172015. View Article : Google Scholar : PubMed/NCBI | |
|
Yang Y, Li M, Yan Y, Zhang J, Sun K, Qu JK, Wang JS and Duan XY: Expression of RAP1B is associated with poor prognosis and promotes an aggressive phenotype in gastric cancer. Oncol Rep. 34:2385–2394. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Heard JJ, Fong V, Bathaie SZ and Tamanoi F: Recent progress in the study of the Rheb family GTPases. Cell Signal. 26:1950–1957. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Armijo ME, Campos T, Fuentes-Villalobos F, Palma ME, Pincheira R and Castro AF: Rheb signaling and tumorigenesis: mTORC1 and new horizons. Int J Cancer. 138:1815–1823. 2016. View Article : Google Scholar | |
|
Basso AD, Mirza A, Liu G, Long BJ, Bishop WR and Kirschmeier P: The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J Biol Chem. 280:31101–31108. 2005. View Article : Google Scholar | |
|
Mecca C, Giambanco I, Donato R and Arcuri C: Targeting mTOR in Glioblastoma: Rationale and Preclinical/Clinical Evidence. Dis Markers. 2018:92304792018. View Article : Google Scholar | |
|
Colardo M, Segatto M and Di Bartolomeo S: Targeting RTK-PI3K-mTOR axis in gliomas: An update. Int J Mol Sci. 22:48992021. View Article : Google Scholar | |
|
Liu M, Bi F, Zhou X and Zheng Y: Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol. 22:365–373. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Hodge RG and Ridley AJ: Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 17:496–510. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
O'Connor K and Chen M: Dynamic functions of RhoA in tumor cell migration and invasion. Small GTPases. 4:141–147. 2013. View Article : Google Scholar | |
|
Yin M, Lu Q, Liu X, Wang T, Liu Y and Chen L: Silencing Drp1 inhibits glioma cells proliferation and invasion by RHOA/ROCK1 pathway. Biochem Biophys Res Commun. 478:663–668. 2016. View Article : Google Scholar | |
|
Xiong W, Yin A, Mao X, Zhang W, Huang H and Zhang X: Resveratrol suppresses human glioblastoma cell migration and invasion via activation of RhoA/ROCK signaling pathway. Oncol Lett. 11:484–490. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Talamillo A, Grande L, Ruiz-Ontañon P, Velasquez C, Mollinedo P, Torices S, Sanchez-Gomez P, Aznar A, Esparis-Ogando A, Lopez-Lopez C, et al: ODZ1 allows glioblastoma to sustain invasiveness through a Myc-dependent transcriptional upregulation of RhoA. Oncogene. 36:1733–1744. 2017. View Article : Google Scholar | |
|
Ridley AJ: Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16:522–529. 2006. View Article : Google Scholar | |
|
Huang M and Prendergast GC: RhoB in cancer suppression. Histol Histopathol. 21:213–218. 2006. | |
|
Zaoui K and Duhamel S: RhoB as a tumor suppressor: It's all about localization. Eur J Cell Biol. 102:1513132023. View Article : Google Scholar : PubMed/NCBI | |
|
Wei LJ, Li JA, Bai DM and Song Y: miR-223-RhoB signaling pathway regulates the proliferation and apoptosis of colon adenocarcinoma. Chem Biol Interact. 289:9–14. 2018. View Article : Google Scholar | |
|
Prendergast GC: Actin' up: RhoB in cancer and apoptosis. Nat Rev Cancer. 1:162–168. 2001. View Article : Google Scholar | |
|
Liu M, Tang Q, Qiu M, Lang N, Li M, Zheng Y and Bi F: miR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett. 585:2998–3005. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Cimbora-Zovko T, Fritz G, Mikac N and Osmak M: Downregulation of RhoB GTPase confers resistance to cisplatin in human laryngeal carcinoma cells. Cancer Lett. 295:182–190. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Kazanietz MG and Caloca MJ: The Rac GTPase in cancer: From old concepts to new paradigms. Cancer Res. 77:5445–5451. 2017. View Article : Google Scholar : | |
|
Steffen A, Ladwein M, Dimchev GA, Hein A, Schwenkmezger L, Arens S, Ladwein KI, Margit Holleboom J, Schur F, Victor Small J, et al: Rac function is crucial for cell migration but is not required for spreading and focal adhesion formation. J Cell Sci. 126:4572–4588. 2013.PubMed/NCBI | |
|
Bosco EE, Mulloy JC and Zheng Y: Rac1 GTPase: A 'Rac' of all trades. Cell Mol Life Sci. 66:370–374. 2009. View Article : Google Scholar | |
|
Leng R, Liao G, Wang H, Kuang J and Tang L: Rac1 expression in epithelial ovarian cancer: effect on cell EMT and clinical outcome. Med Oncol. 32:3292015. View Article : Google Scholar | |
|
Dokmanovic M, Hirsch DS, Shen Y and Wu WJ: Rac1 contributes to trastuzumab resistance of breast cancer cells: Rac1 as a potential therapeutic target for the treatment of trastuzumab-resistant breast cancer. Mol Cancer Ther. 8:1557–1569. 2009. View Article : Google Scholar | |
|
Heid I, Lubeseder-Martellato C, Sipos B, Mazur PK, Lesina M, Schmid RM and Siveke JT: Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 141:719–730. 730.e1–7. 2011. View Article : Google Scholar | |
|
Kwiatkowska A, Didier S, Fortin S, Chuang Y, White T, Berens ME, Rushing E, Eschbacher J, Tran NL, Chan A and Symons M: The small GTPase RhoG mediates glioblastoma cell invasion. Mol Cancer. 11:652012. View Article : Google Scholar : PubMed/NCBI | |
|
Cerione RA: Cdc42: New roads to travel. Trends Cell Biol. 14:127–132. 2004. View Article : Google Scholar | |
|
Etienne-Manneville S: Cdc42-the centre of polarity. J Cell Sci. 117:1291–1300. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Kamai T, Yamanishi T, Shirataki H, Takagi K, Asami H, Ito Y and Yoshida K: Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 10:4799–4805. 2004. View Article : Google Scholar | |
|
Chen QY, Jiao DM, Yao QH, Yan J, Song J, Chen FY, Lu GH and Zhou JY: Expression analysis of Cdc42 in lung cancer and modulation of its expression by curcumin in lung cancer cell lines. Int J Oncol. 40:1561–1568. 2012. | |
|
Zucman-Rossi J, Legoix P and Thomas G: Identification of new members of the Gas2 and Ras families in the 22q12 chromosome region. Genomics. 38:247–254. 1996. View Article : Google Scholar : PubMed/NCBI | |
|
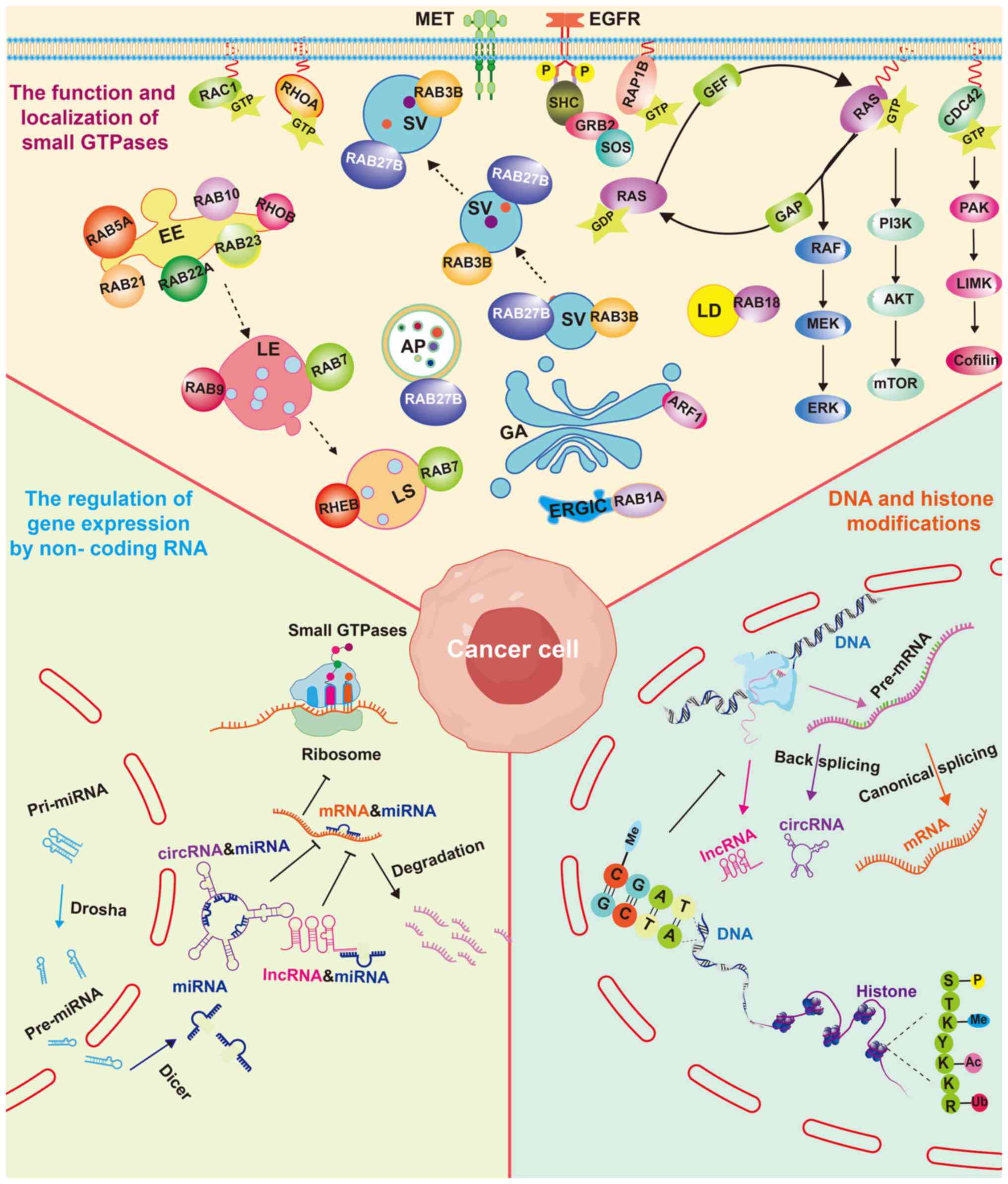
Zhen Y and Stenmark H: Cellular functions of Rab GTPases at a glance. J Cell Sci. 128:3171–3176. 2015. | |
|
Stenmark H: Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009. View Article : Google Scholar | |
|
Homma Y, Hiragi S and Fukuda M: Rab family of small GTPases: An updated view on their regulation and functions. FEBS J. 288:36–55. 2021. View Article : Google Scholar : | |
|
Liu Y, Wang X, Zhang Z, Xiao B, An B and Zhang J: The overexpression of Rab9 promotes tumor progression regulated by XBP1 in breast cancer. Onco Targets Ther. 12:1815–1824. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Li Z, Li Y, Jia Y, Ding B and Yu J: Rab1A knockdown represses proliferation and promotes apoptosis in gastric cancer cells by inhibition of mTOR/p70S6K pathway. Arch Biochem Biophys. 685:1083522020. View Article : Google Scholar | |
|
Zhao Z, Liu XF, Wu HC, Zou SB, Wang JY, Ni PH, Chen XH and Fan QS: Rab5a overexpression promoting ovarian cancer cell proliferation may be associated with APPL1-related epidermal growth factor signaling pathway. Cancer Sci. 101:1454–1462. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang D, Lu C and Ai H: Rab5a is overexpressed in oral cancer and promotes invasion through ERK/MMP signaling. Mol Med Rep. 16:4569–4576. 2017. View Article : Google Scholar | |
|
Ge J and Ge C: Rab14 overexpression regulates gemcitabine sensitivity through regulation of Bcl-2 and mitochondrial function in pancreatic cancer. Virchows Arch. 474:59–69. 2019. View Article : Google Scholar | |
|
Yang XZ, Li XX, Zhang YJ, Rodriguez-Rodriguez L, Xiang MQ, Wang HY and Zheng XF: Rab1 in cell signaling, cancer and other diseases. Oncogene. 35:5699–5704. 2016. View Article : Google Scholar | |
|
Zenner HL, Yoshimura S, Barr FA and Crump CM: Analysis of Rab GTPase-activating proteins indicates that Rab1a/b and Rab43 are important for herpes simplex virus 1 secondary envelopment. J Virol. 85:8012–8021. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Nuoffer C, Davidson HW, Matteson J, Meinkoth J and Balch WE: A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 125:225–237. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang Y, Wang L, Lv Y, Jiang C, Wu G, Dull RO, Minshall RD, Malik AB and Hu G: The GTPase Rab1 is required for NLRP3 inflammasome activation and inflammatory lung injury. J Immunol. 202:194–206. 2019. View Article : Google Scholar | |
|
Yang XZ, Cui SZ, Zeng LS, Cheng TT, Li XX, Chi J, Wang R, Zheng XF and Wang HY: Overexpression of Rab1B and MMP9 predicts poor survival and good response to chemotherapy in patients with colorectal cancer. Aging (Albany NY). 9:914–931. 2017. View Article : Google Scholar | |
|
Wang X, Liu F, Qin X, Huang T, Huang B, Zhang Y and Jiang B: Expression of Rab1A is upregulated in human lung cancer and associated with tumor size and T stage. Aging (Albany NY). 8:2790–2798. 2016. View Article : Google Scholar | |
|
Xu M, Shao X, Kuai X, Zhang L, Zhou C and Cheng Z: Expression analysis and implication of Rab1A in gastrointestinal relevant tumor. Sci Rep. 9:133842019. View Article : Google Scholar : PubMed/NCBI | |
|
Ren B, Wang L, Nan Y, Liu T, Zhao L, Ma H, Li J, Zhang Y and Ren X: RAB1A regulates glioma cellular proliferation and invasion via the mTOR signaling pathway and epithelial-mesenchymal transition. Future Oncol. 17:3203–3216. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Lanzetti L, Palamidessi A, Areces L, Scita G and Di Fiore PP: Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 429:309–314. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Chiariello M, Bruni CB and Bucci C: The small GTPases Rab5a, Rab5b and Rab5c are differentially phosphorylated in vitro. FEBS Lett. 453:20–24. 1999. View Article : Google Scholar : PubMed/NCBI | |
|
Tan JY, Jia LQ, Shi WH, He Q, Zhu L and Yu B: Rab5a-mediated autophagy regulates the phenotype and behavior of vascular smooth muscle cells. Mol Med Rep. 14:4445–4453. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Li Y, Zhao Y, Hu J, Xiao J, Qu L, Wang Z, Ma D and Chen Y: A novel ER-localized transmembrane protein, EMC6, interacts with RAB5A and regulates cell autophagy. Autophagy. 9:150–163. 2013. View Article : Google Scholar | |
|
Yu MH, Luo Y, Qin SL and Zhong M: Increased expression of Rab5A predicts metastasis and poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 8:6974–6980. 2015. | |
|
Engebraaten O, Yau C, Berg K, Borgen E, Garred Ø, Berstad MEB, Fremstedal ASV, DeMichele A, Veer LV', Esserman L and Weyergang A: RAB5A expression is a predictive biomarker for trastuzumab emtansine in breast cancer. Nat Commun. 12:64272021. View Article : Google Scholar | |
|
English AR and Voeltz GK: Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol. 15:169–178. 2013. View Article : Google Scholar | |
|
Chen YT, Holcomb C and Moore HP: Expression and localization of two low molecular weight GTP-binding proteins, Rab8 and Rab10, by epitope tag. Proc Natl Acad Sci USA. 90:6508–6512. 1993. View Article : Google Scholar | |
|
Leaf DS and Blum LD: Analysis of rab10 localization in sea urchin embryonic cells by three-dimensional reconstruction. Exp Cell Res. 243:39–49. 1998. View Article : Google Scholar | |
|
Cardoso CM, Jordao L and Vieira OV: Rab10 regulates phagosome maturation and its overexpression rescues Mycobacterium-containing phagosomes maturation. Traffic. 11:221–235. 2010. View Article : Google Scholar | |
|
Babbey CM, Bacallao RL and Dunn KW: Rab10 associates with primary cilia and the exocyst complex in renal epithelial cells. Am J Physiol Renal Physiol. 299:F495–F506. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Lerner DW, McCoy D, Isabella AJ, Mahowald AP, Gerlach GF, Chaudhry TA and Horne-Badovinac S: A Rab10-dependent mechanism for polarized basement membrane secretion during organ morphogenesis. Dev Cell. 24:159–168. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Isabella AJ and Horne-Badovinac S: Rab10-mediated secretion synergizes with tissue movement to build a polarized basement membrane architecture for organ morphogenesis. Dev Cell. 38:47–60. 2016. View Article : Google Scholar | |
|
Etoh K and Fukuda M: Rab10 regulates tubular endosome formation through KIF13A and KIF13B motors. J Cell Sci. 132:jcs2269772019. View Article : Google Scholar | |
|
Zhuo J, Han J, Zhao Y, Hao R, Shen C, Li H, Dai L, Sheng A, Yao H, Yang X and Liu W: RAB10 promotes breast cancer proliferation migration and invasion predicting a poor prognosis for breast cancer. Sci Rep. 13:152522023. View Article : Google Scholar : | |
|
Wang W, Jia WD, Hu B and Pan YY: RAB10 overexpression promotes tumor growth and indicates poor prognosis of hepatocellular carcinoma. Oncotarget. 8:26434–26447. 2017. View Article : Google Scholar : | |
|
Scita G and Di Fiore PP: The endocytic matrix. Nature. 463:464–473. 2010. View Article : Google Scholar | |
|
Simpson JC and Jones AT: Early endocytic Rabs: Functional prediction to functional characterization. Biochem Soc Symp. 72:99–108. 2005. View Article : Google Scholar | |
|
Simpson JC, Griffiths G, Wessling-Resnick M, Fransen JA, Bennett H and Jones AT: A role for the small GTPase Rab21 in the early endocytic pathway. J Cell Sci. 117(Pt 26): 6297–6311. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Evans TM, Ferguson C, Wainwright BJ, Parton RG and Wicking C: Rab23, a negative regulator of hedgehog signaling, localizes to the plasma membrane and the endocytic pathway. Traffic. 4:869–884. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Pellinen T, Arjonen A, Vuoriluoto K, Kallio K, Fransen JA and Ivaska J: Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of beta1-integrins. J Cell Biol. 173:767–780. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou Y, Wu B, Li JH, Nan G, Jiang JL and Chen ZN: Rab22a enhances CD147 recycling and is required for lung cancer cell migration and invasion. Exp Cell Res. 357:9–16. 2017. View Article : Google Scholar | |
|
Takeda M, Koseki J, Takahashi H, Miyoshi N, Nishida N, Nishimura J, Hata T, Matsuda C, Mizushima T, Yamamoto H, et al: Disruption of Endolysosomal RAB5/7 efficiently eliminates colorectal cancer stem cells. Cancer Res. 79:1426–1437. 2019. View Article : Google Scholar | |
|
Chen Y, Ng F and Tang BL: Rab23 activities and human cancer-emerging connections and mechanisms. Tumour Biol. 37:12959–12967. 2016. View Article : Google Scholar | |
|
Lledo PM, Johannes L, Vernier P, Zorec R, Darchen F, Vincent JD, Henry JP and Mason WT: Rab3 proteins: Key players in the control of exocytosis. Trends Neurosci. 17:426–432. 1994. View Article : Google Scholar | |
|
Martin S and Parton RG: Characterization of Rab18, a lipid droplet-associated small GTPase. Methods Enzymol. 438:109–129. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Gerondopoulos A, Bastos RN, Yoshimura S, Anderson R, Carpanini S, Aligianis I, Handley MT and Barr FA: Rab18 and a Rab18 GEF complex are required for normal ER structure. J Cell Biol. 205:707–720. 2014. View Article : Google Scholar : | |
|
Dejgaard SY, Murshid A, Erman A, Kizilay O, Verbich D, Lodge R, Dejgaard K, Ly-Hartig TB, Pepperkok R, Simpson JC and Presley JF: Rab18 and Rab43 have key roles in ER-Golgi trafficking. J Cell Sci. 121(Pt 16): 2768–2781. 2008. View Article : Google Scholar | |
|
Fukuda M: Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic. 14:949–963. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al: Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 12:19–30. 2010. View Article : Google Scholar | |
|
Li Z, Fang R, Fang J, He S and Liu T: Functional implications of Rab27 GTPases in cancer. Cell Commun Signal. 16:442018. View Article : Google Scholar | |
|
Donaldson JG and Jackson CL: ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 12:362–375. 2011. View Article : Google Scholar | |
|
D'Souza-Schorey C and Chavrier P: ARF proteins: Roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 7:347–358. 2006. View Article : Google Scholar | |
|
Donaldson JG and Honda A: Localization and function of Arf family GTPases. Biochem Soc Trans. 33(Pt 4): 639–642. 2005. View Article : Google Scholar | |
|
Gillingham AK and Munro S: The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 23:579–611. 2007. View Article : Google Scholar | |
|
Schlienger S, Campbell S and Claing A: ARF1 regulates the Rho/MLC pathway to control EGF-dependent breast cancer cell invasion. Mol Biol Cell. 25:17–29. 2014. View Article : Google Scholar | |
|
Boulay PL, Schlienger S, Lewis-Saravalli S, Vitale N, Ferbeyre G and Claing A: ARF1 controls proliferation of breast cancer cells by regulating the retinoblastoma protein. Oncogene. 30:3846–3861. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Ma H, Fang W, Li Q, Wang Y and Hou SX: Arf1 ablation in colorectal cancer cells activates a super signal complex in DC to enhance anti-tumor immunity. Adv Sci (Weinh). 10:e23050892023. View Article : Google Scholar | |
|
Wen PY and Kesari S: Malignant gliomas in adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Rong L, Li N and Zhang Z: Emerging therapies for glioblastoma: Current state and future directions. J Exp Clin Cancer Res. 41:1422022. View Article : Google Scholar : PubMed/NCBI | |
|
Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, Bendszus M, Balana C, Chinot O, Dirven L, et al: EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 18:170–186. 2021. View Article : Google Scholar | |
|
Li T, Li J, Chen Z, Zhang S, Li S, Wageh S, Al-Hartomy OA, Al-Sehemi AG, Xie Z, Kankala RK and Zhang H: Glioma diagnosis and therapy: Current challenges and nanomaterial-based solutions. J Control Release. 352:338–370. 2022. View Article : Google Scholar | |
|
Liu F, Wang Y, Gu H and Wang X: Technologies and applications of single-cell DNA methylation sequencing. Theranostics. 13:2439–2454. 2023. View Article : Google Scholar : | |
|
Alshammari E, Zhang Y, Sobota J and Yang Z: Aberrant DNA methylation of tumor suppressor genes and oncogenes as cancer biomarkers. Genomic and Epigenomic Biomarkers of Toxicology and Disease: Clinical and Therapeutic Actions. Sahu SC: Wiley; Oxford: pp. 251–271. 2022, View Article : Google Scholar | |
|
Da Costa EM, McInnes G, Beaudry A and Raynal NJ: DNA Methylation-targeted drugs. Cancer J. 23:270–276. 2017. View Article : Google Scholar | |
|
Diesch J, Zwick A, Garz AK, Palau A, Buschbeck M and Götze KS: A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin Epigenetics. 8:712016. View Article : Google Scholar : | |
|
Federici L, Capelle L, Annereau M, Bielle F, Willekens C, Dehais C, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Idbaih A, et al: 5-Azacitidine in patients with IDH1/2-mutant recurrent glioma. Neuro Oncol. 22:1226–1228. 2020. View Article : Google Scholar | |
|
He W, Lin S, Guo Y, Wu Y, Zhang LL, Deng Q, Du ZM, Wei M, Zhu W, Chen WJ, et al: Targeted demethylation at ZNF154 promotor upregulates ZNF154 expression and inhibits the proliferation and migration of Esophageal squamous carcinoma cells. Oncogene. 41:4537–4546. 2022. View Article : Google Scholar | |
|
Zaib S, Rana N and Khan I: Histone modifications and their role in epigenetics of cancer. Curr Med Chem. 29:2399–2411. 2022. View Article : Google Scholar | |
|
Chen R, Zhang M, Zhou Y, Guo W, Yi M, Zhang Z, Ding Y and Wang Y: The application of histone deacetylases inhibitors in glioblastoma. J Exp Clin Cancer Res. 39:1382020. View Article : Google Scholar : PubMed/NCBI | |
|
Wang F, Jin Y, Wang M, Luo HY, Fang WJ, Wang YN, Chen YX, Huang RJ, Guan WL, Li JB, et al: Combined anti-PD-1, HDAC inhibitor and anti-VEGF for MSS/pMMR colorectal cancer: A randomized phase 2 trial. Nat Med. 30:1035–1043. 2024. View Article : Google Scholar : PubMed/NCBI | |
|
Gao Y, He H, Li X, Zhang L, Xu W, Feng R, Li W, Xiao Y, Liu X, Chen Y, et al: Sintilimab (anti-PD-1 antibody) plus chidamide (histone deacetylase inhibitor) in relapsed or refractory extranodal natural killer T-cell lymphoma (SCENT): A phase Ib/II study. Signal Transduct Target Ther. 9:1212024. View Article : Google Scholar : | |
|
Monje M, Cooney T, Glod J, Huang J, Peer CJ, Faury D, Baxter P, Kramer K, Lenzen A, Robison NJ, et al: Phase I trial of panobinostat in children with diffuse intrinsic pontine glioma: A report from the Pediatric brain tumor consortium (PBTC-047). Neuro Oncol. 25:2262–2272. 2023. View Article : Google Scholar : | |
|
Mueller S, Kline C, Stoller S, Lundy S, Christopher L, Reddy AT, Banerjee A, Cooney TM, Raber S, Hoffman C, et al: PNOC015: Repeated convection-enhanced delivery of MTX110 (aqueous panobinostat) in children with newly diagnosed diffuse intrinsic pontine glioma. Neuro Oncol. 25:2074–2086. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Xu K, Ramesh K, Huang V, Gurbani SS, Cordova JS, Schreibmann E, Weinberg BD, Sengupta S, Voloschin AD, Holdhoff M, et al: Final report on clinical outcomes and tumor recurrence patterns of a pilot study assessing efficacy of belinostat (PXD-101) with chemoradiation for newly diagnosed glioblastoma. Tomography. 8:688–700. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Raedler LA: Farydak (Panobinostat): First HDAC inhibitor approved for patients with relapsed multiple myeloma. Am Health Drug Benefits. 9(Spec Feature): 84–87. 2016.PubMed/NCBI | |
|
Zhang L, Liao Y and Tang L: MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res. 38:532019. View Article : Google Scholar : | |
|
Li Z and Rana TM: Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 13:622–638. 2014. View Article : Google Scholar | |
|
Petrescu GED, Sabo AA, Torsin LI, Calin GA and Dragomir MP: MicroRNA based theranostics for brain cancer: Basic principles. J Exp Clin Cancer Res. 38:2312019. View Article : Google Scholar : PubMed/NCBI | |
|
Mathupala SP, Mittal S, Guthikonda M and Sloan AE: MicroRNA and brain tumors: A cause and a cure? DNA Cell Biol. 26:301–310. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Morokoff A, Jones J, Nguyen H, Ma C, Lasocki A, Gaillard F, Bennett I, Luwor R, Stylli S, Paradiso L, et al: Serum microRNA is a biomarker for post-operative monitoring in glioma. J Neurooncol. 149:391–400. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Yue X, Lan F, Hu M, Pan Q, Wang Q and Wang J: Downregulation of serum microRNA-205 as a potential diagnostic and prognostic biomarker for human glioma. J Neurosurg. 124:122–128. 2016. View Article : Google Scholar | |
|
Sun Y, Jing Y and Zhang Y: Serum lncRNA-ANRIL and SOX9 expression levels in glioma patients and their relationship with poor prognosis. World J Surg Oncol. 19:2872021. View Article : Google Scholar : | |
|
Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C and Ayad NG: Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. 17:742018. View Article : Google Scholar : | |
|
Stella M, Falzone L, Caponnetto A, Gattuso G, Barbagallo C, Battaglia R, Mirabella F, Broggi G, Altieri R, Certo F, et al: Serum extracellular vesicle-derived circHIPK3 and circS-MARCA5 are two novel diagnostic biomarkers for glioblastoma multiforme. Pharmaceuticals (Basel). 14:6182021. View Article : Google Scholar | |
|
Li P, Xu Z, Liu T, Liu Q, Zhou H, Meng S, Feng Z, Tang Y, Liu C, Feng J, et al: Circular RNA sequencing reveals serum exosome circular RNA panel for high-grade astrocytoma diagnosis. Clin Chem. 68:332–343. 2022. View Article : Google Scholar |










