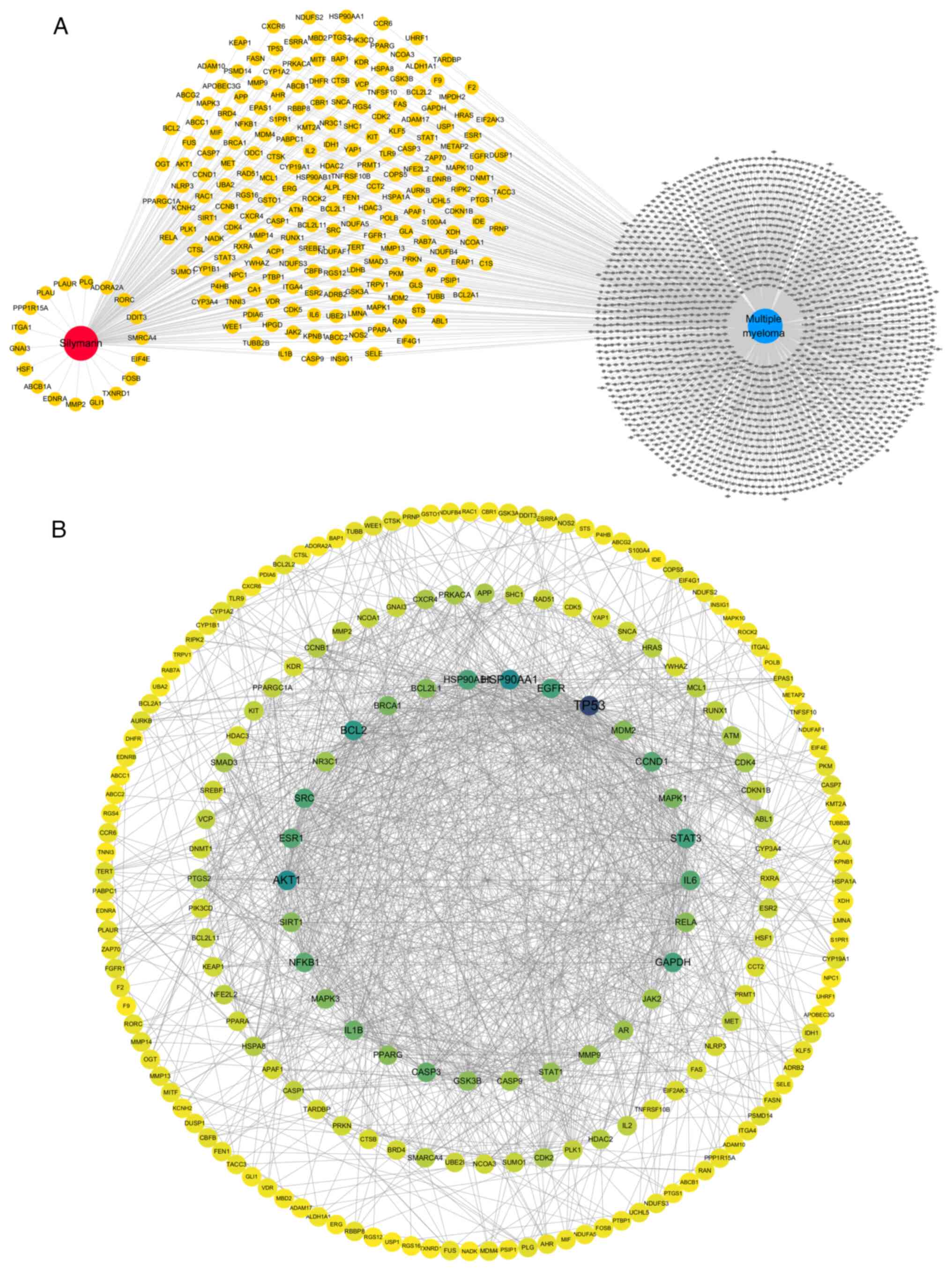
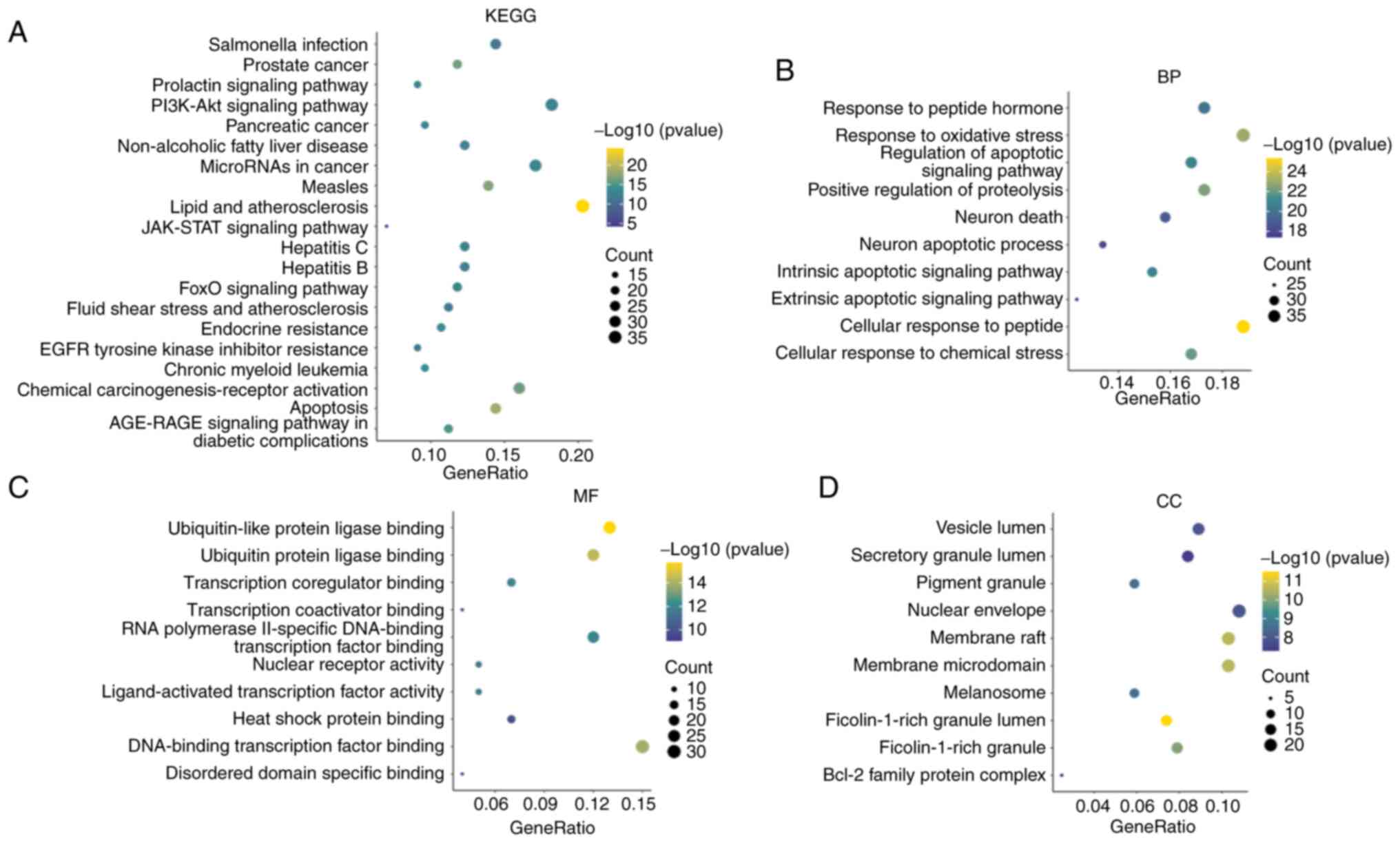
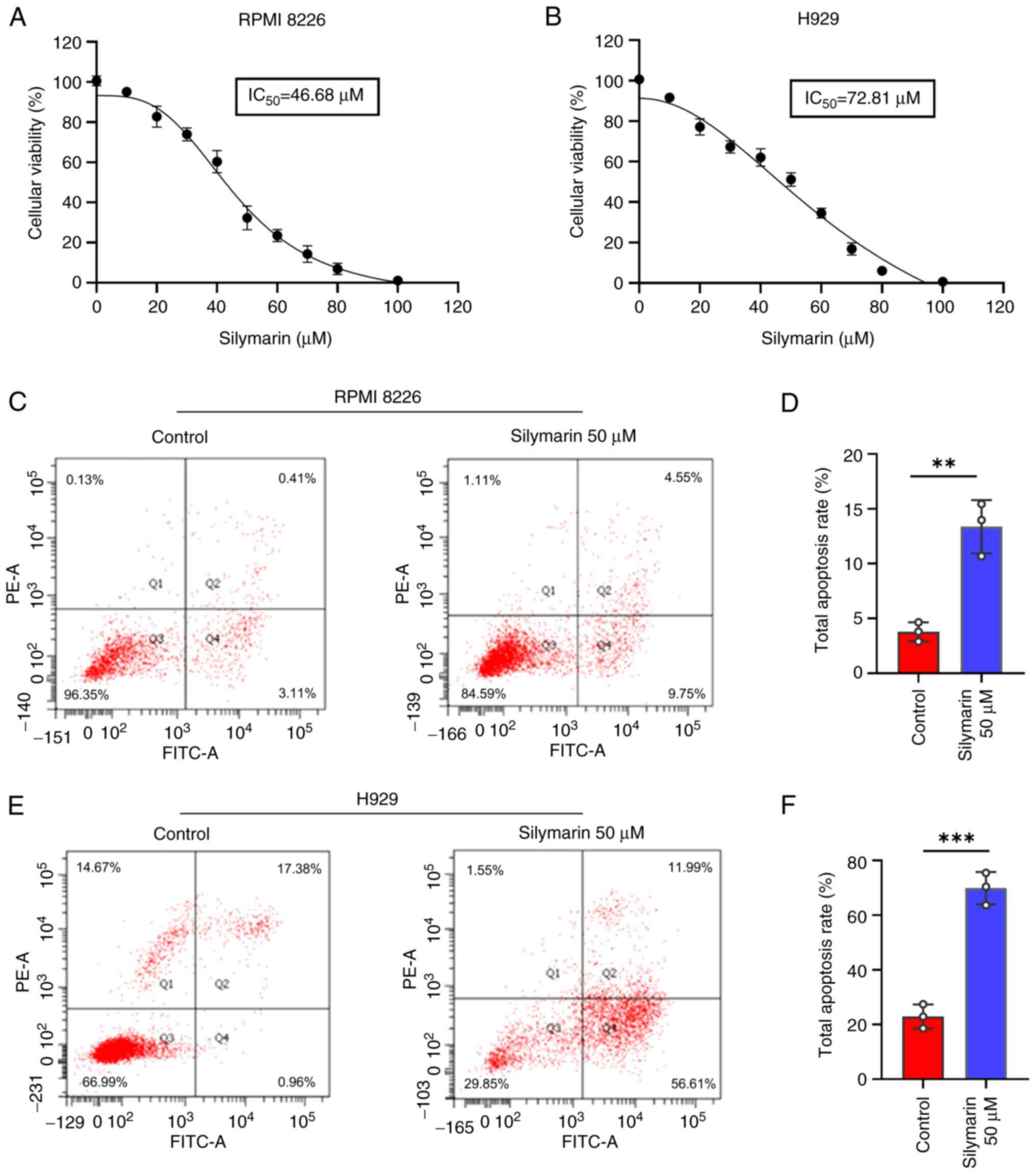
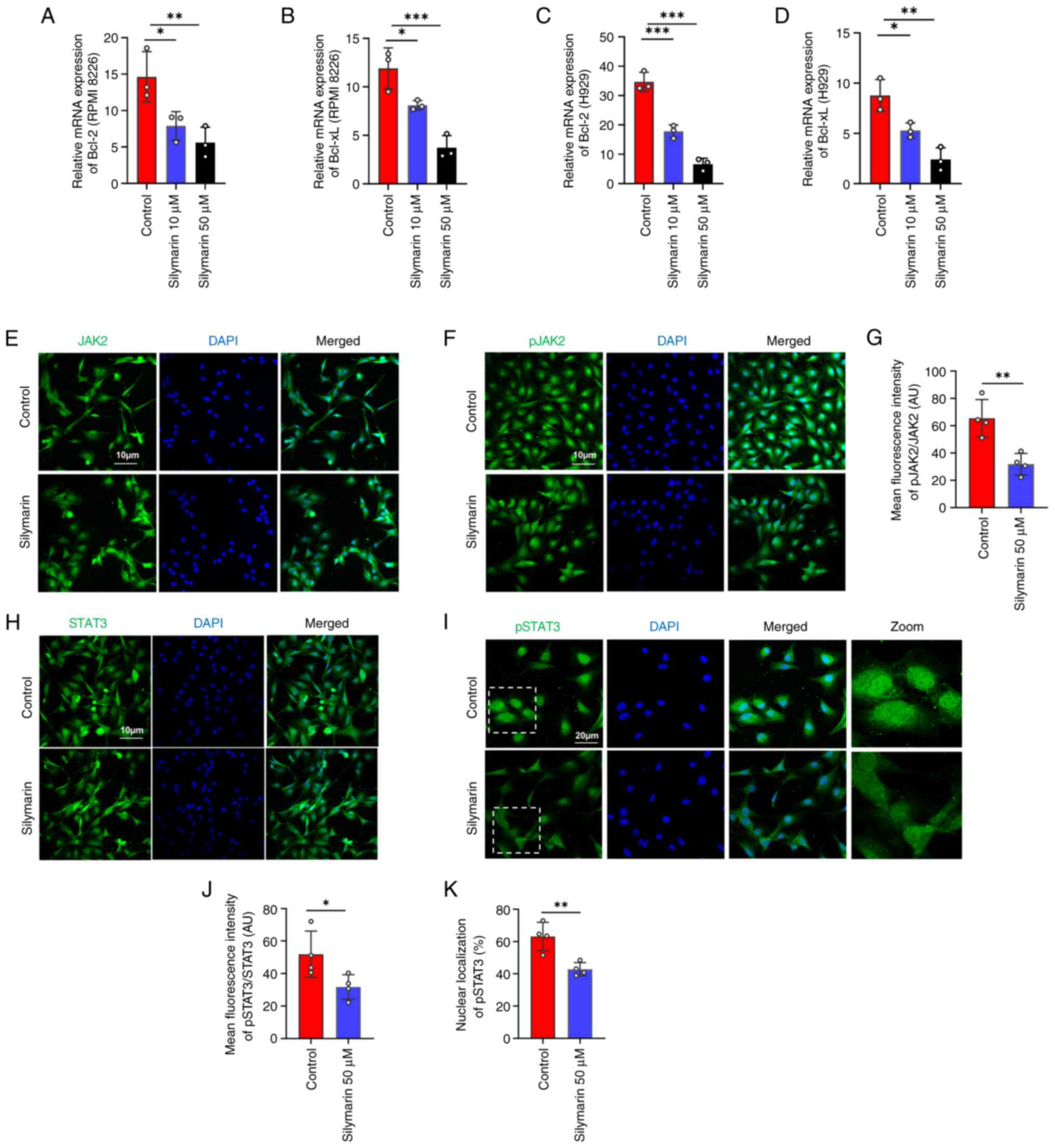
|
1
|
Raab MS, Podar K, Breitkreutz I,
Richardson PG and Anderson KC: Multiple myeloma. Lancet.
374:324–339. 2009. View Article : Google Scholar
|
|
2
|
Cowan AJ, Green DJ, Kwok M, Lee S, Coffey
DG, Holmberg LA, Tuazon S, Gopal AK and Libby EN: Diagnosis and
management of multiple myeloma: A review. JAMA. 327:464–477. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choi T, Ahn W, Shin DW, Han K, Kim D and
Chun S: Association between kidney function, proteinuria and the
risk of multiple myeloma: A population-based retrospective cohort
study in South Korea. Cancer Res Treat. 54:926–936. 2022.
View Article : Google Scholar
|
|
4
|
Korbet SM and Schwartz MM: Multiple
myeloma. J Am Soc Nephrol. 17:2533–2545. 2006. View Article : Google Scholar
|
|
5
|
Rajkumar SV: Myeloma today: Disease
definitions and treatment advances. Am J Hematol. 91:90–100. 2016.
View Article : Google Scholar
|
|
6
|
Turesson I, Bjorkholm M, Blimark CH,
Kristinsson S, Velez R and Landgren O: Rapidly changing myeloma
epidemiology in the general population: Increased incidence, older
patients, and longer survival. Eur J Haematol. April 9–2018.(Epub
ahead of print). View Article : Google Scholar
|
|
7
|
Malard F, Neri P, Bahlis NJ, Terpos E,
Moukalled N, Hungria VTM, Manier S and Mohty M: Multiple myeloma.
Nat Rev Dis Primers. 10:452024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng Y, Tu Y, Lao S, Wu M, Yin H, Wang L
and Liao W: The role and mechanism of citrus flavonoids in
cardiovascular diseases prevention and treatment. Crit Rev Food Sci
Nutr. 62:7591–7614. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Majee C, Mazumder R and Choudhary AN:
Salahuddin: An insight into the hepatoprotective activity and
structure-activity relationships of flavonoids. Mini Rev Med Chem.
23:131–149. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mohammadi S, Ashtary-Larky D, Asbaghi O,
Farrokhi V, Jadidi Y, Mofidi F, Mohammadian M and Afrisham R:
Effects of silymarin supplementation on liver and kidney functions:
A systematic review and dose-response meta-analysis. Phytother Res.
38:2572–2593. 2024. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gillessen A and Schmidt HH: Silymarin as
supportive treatment in liver diseases: A narrative review. Adv
Ther. 37:1279–1301. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wadhwa K, Pahwa R, Kumar M, Kumar S,
Sharma PC, Singh G, Verma R, Mittal V, Singh I, Kaushik D and
Jeandet P: Mechanistic insights into the pharmacological
significance of silymarin. Molecules. 27:53272022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Delmas D, Xiao J, Vejux A and Aires V:
Silymarin and cancer: A dual strategy in both in chemoprevention
and chemosensitivity. Molecules. 25:20092020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koltai T and Fliegel L: Role of silymarin
in cancer treatment: Facts, hypotheses, and questions. J Evid Based
Integr Med. 27:2515690X2110688262022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng N, Luo J and Guo X: Silybin
suppresses cell proliferation and induces apoptosis of multiple
myeloma cells via the PI3K/Akt/mTOR signaling pathway. Mol Med Rep.
13:3243–3248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
UniProt Consortium: UniProt: The universal
protein knowledgebase in 2025. Nucleic Acids Res. 53(D1):
D609–D617. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carvalho-Silva D, Pierleoni A, Pignatelli
M, Ong C, Fumis L, Karamanis N, Carmona M, Faulconbridge A,
Hercules A, McAuley E, et al: Open targets platform: New
developments and updates two years on. Nucleic Acids Res. 47(D1):
D1056–D1065. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piñero J, Saüch J, Sanz F and Furlong LI:
The DisGeNET cytoscape app: Exploring and visualizing disease
genomics data. Comput Struct Biotechnol J. 19:2960–2967. 2021.
View Article : Google Scholar
|
|
19
|
Davis AP, Grondin CJ, Johnson RJ, Sciaky
D, McMorran R, Wiegers J, Wiegers TC and Mattingly CJ: The
comparative toxicogenomics database: Update 2019. Nucleic Acids
Res. 47(D1): D948–D954. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Safran M, Dalah I, Alexander J, Rosen N,
Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al:
GeneCards version 3: The human gene integrator. Database (Oxford).
2010:baq0202010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karbalaei R, Allahyari M, Rezaei-Tavirani
M, Asadzadeh-Aghdaei H and Zali MR: Protein-protein interaction
analysis of Alzheimer's disease and NAFLD based on systems biology
methods unhide common ancestor pathways. Gastroenterol Hepatol Bed
Bench. 11:27–33. 2018.PubMed/NCBI
|
|
25
|
Assenov Y, Ramírez F, Schelhorn SE,
Lengauer T and Albrecht M: Computing topological parameters of
biological networks. Bioinformatics. 24:282–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim S, Thiessen PA, Bolton EE, Chen J, Fu
G, Gindulyte A, Han L, He J, He S, Shoemaker BA, et al: PubChem
substance and compound databases. Nucleic Acids Res. 44(D1):
D1202–D1213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Boyle NM, Banck M, James CA, Morley C,
Vandermeersch T and Hutchison GR: Open Babel: An open chemical
toolbox. J Cheminform. 3:332011. View Article : Google Scholar
|
|
30
|
Karuppasamy MP, Venkateswaran S and
Subbiah P: PDB-2-PBv3.0: An updated protein block database. J
Bioinform Comput Biol. 18:20500092020. View Article : Google Scholar
|
|
31
|
Li B, Rui J, Ding X and Yang X: Exploring
the multicomponent synergy mechanism of Banxia Xiexin Decoction on
irritable bowel syndrome by a systems pharmacology strategy. J
Ethnopharmacol. 233:158–168. 2019. View Article : Google Scholar
|
|
32
|
Li X, Xu X, Wang J, Yu H, Wang X, Yang H,
Xu H, Tang S, Li Y, Yang L, et al: A system-level investigation
into the mechanisms of Chinese traditional medicine: Compound
danshen formula for cardiovascular disease treatment. PLoS One.
7:e439182012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang P, Zhang D, Zhou W, Wang L, Wang B,
Zhang T and Li S: Network pharmacology: Towards the artificial
intelligence-based precision traditional Chinese medicine. Brief
Bioinform. 25:bbad5182023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu X, Li J, Fu M, Zhao X and Wang W: The
JAK/STAT signaling pathway: From bench to clinic. Signal Transduct
Target Ther. 6:4022021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fruman DA and Rommel C: PI3K and cancer:
Lessons, challenges and opportunities. Nat Rev Drug Discov.
13:140–156. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ma J, Matkar S, He X and Hua X: FOXO
family in regulating cancer and metabolism. Semin Cancer Biol.
50:32–41. 2018. View Article : Google Scholar
|
|
37
|
Bosch-Barrera J and Menendez JA: Silibinin
and STAT3: A natural way of targeting transcription factors for
cancer therapy. Cancer Treat Rev. 41:540–546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liakopoulou C, Kazazis C and Vallianou NG:
Silimarin and cancer. Anticancer Agents Med Chem. 18:1970–1974.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim SH, Choo GS, Yoo ES, Woo JS, Lee JH,
Han SH, Jung SH, Kim HJ and Jung JY: Silymarin inhibits
proliferation of human breast cancer cells via regulation of the
MAPK signaling pathway and induction of apoptosis. Oncol Lett.
21:4922021. View Article : Google Scholar
|
|
40
|
Yu L, Li T, Zhang H, Ma Z and Wu S:
Silymarin suppresses proliferation of human hepatocellular
carcinoma cells under hypoxia through downregulation of the
HIF-1α/VEGF pathway. Am J Transl Res. 15:4521–4532. 2023.PubMed/NCBI
|
|
41
|
Borensztejn A, Boissoneau E, Fernandez G,
Agnès F and Pret AM: JAK/STAT autocontrol of ligand-producing cell
number through apoptosis. Development. 140:195–204. 2013.
View Article : Google Scholar
|
|
42
|
Weber-Nordt RM, Mertelsmann R and Finke J:
The JAK-STAT pathway: signal transduction involved in
proliferation, differentiation and transformation. Leuk Lymphoma.
28:459–467. 1998. View Article : Google Scholar
|
|
43
|
Bharti AC, Shishodia S, Reuben JM, Weber
D, Alexanian R, Raj-Vadhan S, Estrov Z, Talpaz M and Aggarwal BB:
Nuclear factor-kappaB and STAT3 are constitutively active in CD138+
cells derived from multiple myeloma patients, and suppression of
these transcription factors leads to apoptosis. Blood.
103:3175–3184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Corvinus FM, Orth C, Moriggl R, Tsareva
SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal
K, et al: Persistent STAT3 activation in colon cancer is associated
with enhanced cell proliferation and tumor growth. Neoplasia.
7:545–555. 2005. View Article : Google Scholar
|
|
45
|
Hodge DR, Hurt EM and Farrar WL: The role
of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer.
41:2502–2512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi Z, Zhou Q, Gao S, Li W, Li X, Liu Z,
Jin P and Jiang J: Silibinin inhibits endometrial carcinoma via
blocking pathways of STAT3 activation and SREBP1-mediated lipid
accumulation. Life Sci. 217:70–80. 2019. View Article : Google Scholar
|
|
47
|
Soleimani V, Delghandi PS, Moallem SA and
Karimi G: Safety and toxicity of silymarin, the major constituent
of milk thistle extract: An updated review. Phytother Res.
33:1627–1638. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yao ZJ, Dong J, Che YJ, Zhu MF, Wen M,
Wang NN, Wang S, Lu AP and Cao DS: TargetNet: A web service for
predicting potential drug-target interaction profiling via
multi-target SAR models. J Comput Aided Mol Des. 30:413–424. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gfeller D, Grosdidier A, Wirth M, Daina A,
Michielin O and Zoete V: SwissTargetPrediction: A web server for
target prediction of bioactive small molecules. Nucleic Acids Res.
42((Web Server Issue)): W32–W38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bosc N, Atkinson F, Felix E, Gaulton A,
Hersey A and Leach AR: Large scale comparison of QSAR and conformal
prediction methods and their applications in drug discovery. J
Cheminform. 11:42019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H,
Diao L, Gu J, Wang W, Li D and He F: BATMAN-TCM: A bioinformatics
analysis tool for molecular mechANism of traditional Chinese
medicine. Sci Rep. 6:211462016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Szklarczyk D, Santos A, von Mering C,
Jensen LJ, Bork P and Kuhn M: STITCH 5: Augmenting protein-chemical
interaction networks with tissue and affinity data. Nucleic Acids
Res. 44(D1): D380–D384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wishart DS, Feunang YD, Guo AC, Lo EJ,
Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al:
DrugBank 5.0: A major update to the DrugBank database for 2018.
Nucleic Acids Res. 46(D1): D1074–D1082. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Y, Zhang S, Li F, Zhou Y, Zhang Y,
Wang Z, Zhang R, Zhu J, Ren Y, Tan Y, et al: Therapeutic target
database 2020: Enriched resource for facilitating research and
early development of targeted therapeutics. Nucleic Acids Res.
48(D1): D1031–D1041. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gaulton A, Hersey A, Nowotka M, Bento AP,
Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ,
Cibrián-Uhalte E, et al: The ChEMBL database in 2017. Nucleic Acids
Res. 45(D1): D945–D954. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Davis AP, Wiegers TC, Johnson RJ, Sciaky
D, Wiegers J and Mattingly CJ: Comparative toxicogenomics database
(CTD): Update 2023. Nucleic Acids Res. 51(D1): D1257–D1262. 2023.
View Article : Google Scholar : PubMed/NCBI
|