|
1
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:6356482020.
View Article : Google Scholar
|
|
2
|
Guan WL, He Y and Xu RH: Gastric cancer
treatment: Recent progress and future perspectives. J Hematol
Oncol. 16:572023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Joshi SS and Badgwell BD: Current
treatment and recent progress in gastric cancer. CA Cancer J Clin.
71:2642792021.
|
|
4
|
Dicken BJ, Bigam DL, Cass C, Mackey JR,
Joy AA and Hamilton SM: Gastric adenocarcinoma: Review and
considerations for future directions. Ann Surg. 241:27392005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ishida S, Andreux P, Poitry-Yamate C,
Auwerx J and Hanahan D: Bioavailable copper modulates oxidative
phosphorylation and growth of tumors. Proc Natl Acad Sci USA.
110:19507–19512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsvetkov P, Coy S, Petrova B, Dreishpoon
M, Verma A, Abdusamad M, Rossen J, JoeschCohen L, Humeidi R,
Spangler RD, et al: Copper induces cell death by targeting
lipoylated TCA cycle proteins. Science. 375:125412612022.
View Article : Google Scholar
|
|
7
|
Cobine PA and Brady DC: Cuproptosis:
Cellular and molecular mechanisms underlying copper-induced cell
death. Mol Cell. 82:1786–1787. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Chen Y, Zhang J, Yang Y, Fleishman
JS, Wang Y, Wang J, Chen J, Li Y and Wang H: Cuproptosis: A novel
therapeutic target for overcoming cancer drug resistance. Drug
Resist Updat. 72:1010182024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen G, Luo D, Qi X, Li D, Zheng J, Luo Y,
Zhang C, Ren Q, Lu Y, Chan YT, et al: Characterization of
cuproptosis in gastric cancer and relationship with clinical and
drug reactions. Front Cell Dev Biol. 11:11728952023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chong W, Ren H, Chen H, Xu K, Zhu X, Liu
Y, Sang Y, Li H, Liu J, Ye C, et al: Clinical features and
molecular landscape of cuproptosis signature-related molecular
subtype in gastric cancer. Imeta. 3:e1902024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Braymer JJ, Freibert SA, RakwalskaBange M
and Lill R: Mechanistic concepts of ironsulfur protein biogenesis
in biology. Biochim Biophys Acta Mol Cell Res. 1868:1188632021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Srinivasan V, Netz DJ, Webert H,
Mascarenhas J, Pierik AJ, Michel H and Lill R: Structure of the
yeast WD40 domain protein Cia1, a component acting late in
ironsulfur protein biogenesis. Structure. 15:124612572007.
View Article : Google Scholar
|
|
13
|
Gari K, León Ortiz AM, Borel V, Flynn H,
Skehel JM and Boulton SJ: MMS19 links cytoplasmic ironsulfur
cluster assembly to DNA metabolism. Science. 337:2432452012.
View Article : Google Scholar
|
|
14
|
Stehling O, Mascarenhas J, Vashisht AA,
Sheftel AD, Niggemeyer B, Rösser R, Pierik AJ, Wohlschlegel JA and
Lill R: Human CIA2AFAM96A and CIA2BFAM96B integrate iron
homeostasis and maturation of different subsets of cytosolicnuclear
ironsulfur proteins. Cell Metab. 18:1871982013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNAsequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez J-C and Müller M: pROC: an open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Therneau T: A package for survival
analysis in R. R package version 2. 2014.
|
|
18
|
Harrell FE: Regression modeling
strategies. With Applications to Linear Models, Logistic
Regression, and Survival Analysis. 1st edition. Springer; New York,
NY: 2001
|
|
19
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. Omics. 16:2842872012. View Article : Google Scholar
|
|
20
|
Stuart T, Butler A, Hoffman P, Hafemeister
C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P and
Satija R: Comprehensive integration of singlecell data. Cell.
177:18881902.e212019. View Article : Google Scholar
|
|
21
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:174717562018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reinhold WC, Sunshine M, Liu H, Varma S,
Kohn KW, Morris J, Doroshow J and Pommier Y: CellMiner: A webbased
suite of genomic and pharmacologic tools to explore transcript and
drug patterns in the NCI60 cell line set. Cancer Res.
72:349935112012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Villanueva RAM and Chen ZJ: ggplot2:
Elegant Graphics for Data Analysis (2nd ed.). Measurement (Mahwah N
J). 17:160–167. 2019.
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using realtime quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:4024082001. View Article : Google Scholar
|
|
25
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: Procedure and some applications. Proc Natl
Acad Sci USA. 76:435043541979. View Article : Google Scholar
|
|
26
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:5505532002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mosmann T: Rapid colorimetric assay for
cellular growth and survival: Application to proliferation and
cytotoxicity assays. J Immunol Methods. 65:55631983. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Franken NA, Rodermond HM, Stap J, Haveman
J and van Bree C: Clonogenic assay of cells in vitro. Nat Protoc.
1:231523192006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Justus CR, Leffler N, RuizEchevarria M and
Yang LV: In vitro cell migration and invasion assays. J Vis Exp.
510462014.PubMed/NCBI
|
|
30
|
Pawley JB: Handbook of biological confocal
microscopy. 3rd edition. Springer Science & Business Media;
Berlin: 2006, View Article : Google Scholar
|
|
31
|
Shen W, Song Z, Zhong X, Huang M, Shen D,
Gao P, Qian X, Wang M, He X, Wang T, et al: Sangerbox: A
comprehensive, interaction-friendly clinical bioinformatics
analysis platform. Imeta. 1:e362022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sipos K, Lange H, Fekete Z, Ullmann P,
Lill R and Kispal G: Maturation of cytosolic iron-sulfur proteins
requires glutathione. J Biol Chem. 277:26944–26949. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Morimoto RI: The heat shock response:
Systems biology of proteotoxic stress in aging and disease. Cold
Spring Harb Symp Quant Biol. 76:91–99. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lianos GD, Alexiou GA, Mangano A, Mangano
A, Rausei S, Boni L, Dionigi G and Roukos DH: The role of heat
shock proteins in cancer. Cancer Lett. 360:1141182015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao Y, Bai Y, Shen M and Li Y:
Therapeutic strategies for gastric cancer targeting immune cells:
Future directions. Front Immunol. 13:9927622022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shitara K, Özgüroğlu M, Bang YJ, Di
Bartolomeo M, Mandalà M, Ryu MH, Fornaro L, Olesiński T, Caglevic
C, Chung HC, et al: Pembrolizumab versus paclitaxel for previously
treated, advanced gastric or gastrooesophageal junction cancer
(KEYNOTE061): A randomised, openlabel, controlled, phase 3 trial.
Lancet. 392:1231332018. View Article : Google Scholar
|
|
37
|
Sexton RE, Al Hallak MN, Diab M and Azmi
AS: Gastric cancer: A comprehensive review of current and future
treatment strategies. Cancer Metastasis Rev. 39:117912032020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bai R, Chen N, Li L, Du N, Bai L, Lv Z,
Tian H and Cui J: Mechanisms of cancer resistance to immunotherapy.
Front Oncol. 10:12902020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Petrillo A, Pompella L, Tirino G,
Pappalardo A, Laterza MM, Caterino M, Orditura M, Ciardiello F,
Lieto E, Galizia G, et al: Perioperative treatment in resectable
gastric cancer: Current perspectives and future directions. Cancers
(Basel). 11:3992019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yuan X, Wang J, Huang Y, Shangguan D and
Zhang P: Singlecell profiling to explore immunological
heterogeneity of tumor microenvironment in breast cancer. Front
Immunol. 12:6436922021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Z, Shao S, Luo H, Sun W, Wang J and
Yin H: The functions of cuproptosis in gastric cancer: Therapy,
diagnosis, prognosis. Biomed Pharmacother. 177:1171002024.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:eaan25072017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lill R, Dutkiewicz R, Elsässer HP,
Hausmann A, Netz DJ, Pierik AJ, Stehling O, Urzica E and Mühlenhoff
U: Mechanisms of iron-sulfur protein maturation in mitochondria,
cytosol and nucleus of eukaryotes. Biochim Biophys Acta.
1763:6526672006.PubMed/NCBI
|
|
44
|
Maio N, Orbach R, Zaharieva IT, Töpf A,
Donkervoort S, Munot P, Mueller J, Willis T, Verma S, Peric S, et
al: CIAO1 loss of function causes a neuromuscular disorder with
compromise of nucleocytoplasmic Fe-S enzymes. J Clin Invest.
134:e1795592024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Johnstone RW, Wang J, Tommerup N, Vissing
H, Roberts T and Shi Y: Ciao 1 is a novel WD40 protein that
interacts with the tumor suppressor protein WT1. J Biol Chem.
273:10880108871998. View Article : Google Scholar
|
|
46
|
Netz DJ, Stith CM, Stümpfig M, Köpf G,
Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM and Pierik AJ:
Eukaryotic DNA polymerases require an ironsulfur cluster for the
formation of active complexes. Nat Chem Biol. 8:1251322012.
View Article : Google Scholar
|
|
47
|
Isaya G: Mitochondrial ironsulfur cluster
dysfunction in neurodegenerative disease. Front Pharmacol.
5:292014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shah NP, Tran C, Lee FY, Chen P, Norris D
and Sawyers CL: Overriding imatinib resistance with a novel ABL
kinase inhibitor. Science. 305:399–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Talpaz M, Shah NP, Kantarjian H, Donato N,
Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E,
et al: Dasatinib in imatinib-resistant Philadelphia
chromosome-positive leukemias. N Engl J Med. 354:2531–2541. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dawson MA, Curry JE, Barber K, Beer PA,
Graham B, Lyons JF, Richardson CJ, Scott MA, Smyth T, Squires MS,
et al: AT9283, a potent inhibitor of the aurora kinases and Jak2,
has therapeutic potential in myeloproliferative disorders. Br J
Haematol. 150:46–57. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Deng J, Zhuang H, Shao S, Zeng X, Xue P,
Bai T, Wang X, Shangguan S, Chen Y, Yan S and Huang W:
Mitochondrial-targeted copper delivery for cuproptosis-based
synergistic cancer therapy. Adv Healthc Mater. 13:e23045222024.
View Article : Google Scholar : PubMed/NCBI
|



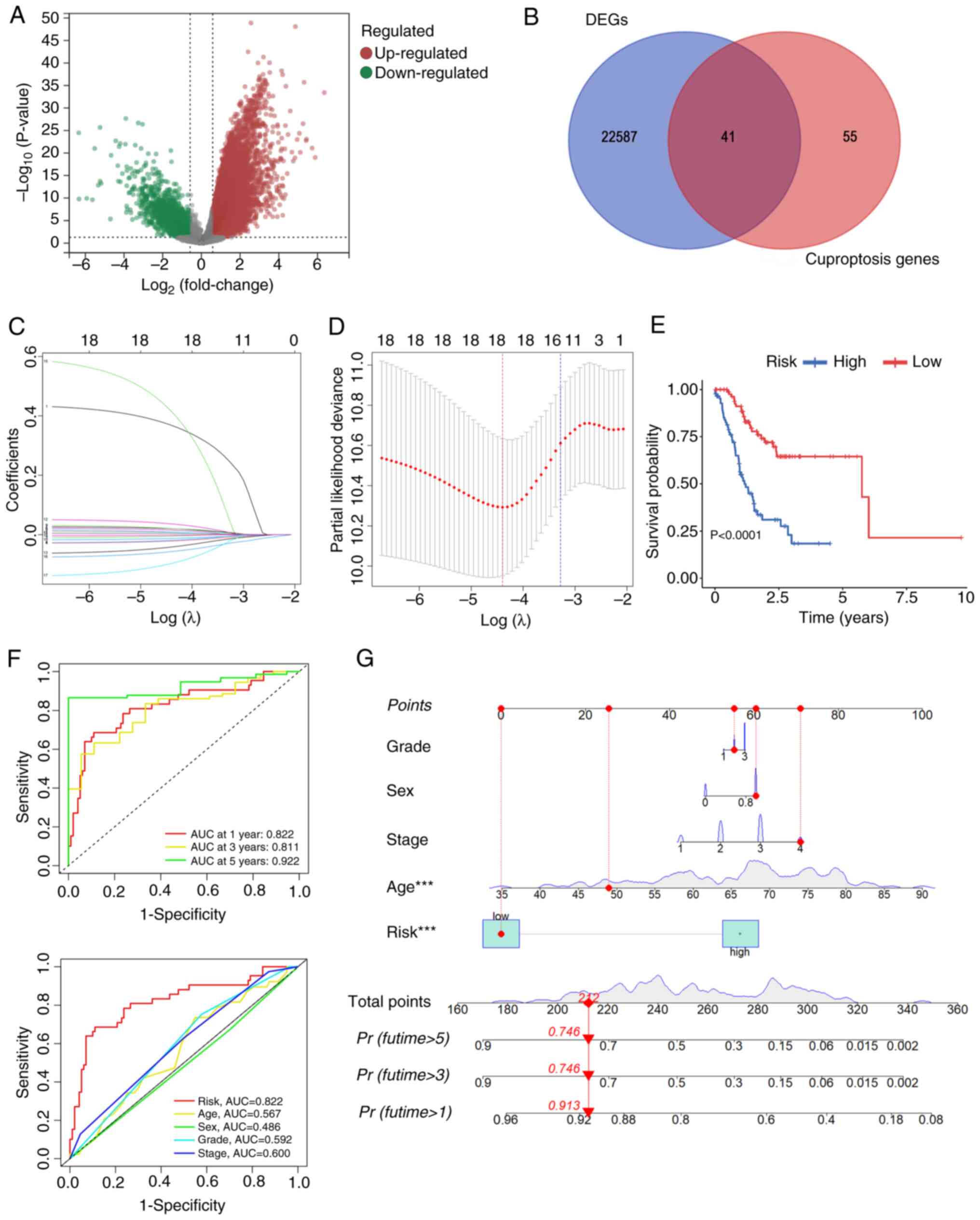
![Enrichment analysis of DEGs and
CIAO1-interacting proteins in stomach adenocarcinoma. (A) Volcano
plot, highlighting the DEGs between the high- and low-CIAO1
expression groups [Log(FoldChange)<1; P<0.05]. (B)
Gene Ontology functional enrichment analysis for DEGs. (C) Geneset
enrichment analysis. (D) Protein-protein interaction network plot
of CIAO1-interacting proteins. (E) Kyoto Encyclopedia of Genes and
Genomes functional enrichment analysis for DEGs. The
three-dimensional visualization of the structure of CIAO1 and its
interacting proteins: (F) CIAO2A, (G) CIAO2B and (H) CIAO3. DEGs,
differentially expressed genes; CIAO, cytosolic iron-sulfur
assembly component; BP, Biological Process; CC, Cellular
Compartment; MF, Molecular Function.](/article_images/ol/30/3/ol-30-03-15186-g06.jpg)