Effects of interleukin‑6 genetic variation on hepatitis B virus infection and susceptibility to hepatocellular carcinoma: A systematic review and meta‑analysis
- Authors:
- Published online on: May 12, 2025 https://doi.org/10.3892/wasj.2025.351
- Article Number: 63
-
Copyright : © Antony et al. This is an open access article distributed under the terms of Creative Commons Attribution License [CC BY 4.0].
Abstract
Introduction
The hepatitis B virus (HBV) is a vast global public health concern as it causes a number of liver-related illnesses, such as chronic hepatitis, cirrhosis and hepatocellular carcinoma (HCC) (1). Among 257 million individuals, only 36 million individuals had chronic hepatitis B, as per a diagnosis made in 2024. This issue has become a main concern for public health across the globe (2). A few adults who are infected with HBV will develop chronic hepatitis B and 20-30% of them would develop liver cancer and/or HCC, which is responsible for an estimated 887,000 related annually (3). Of note, >80 million individuals and 37,000 children in China are chronically infected with HBV; this renders China as the country with the highest burden of infection despite the availability of effective vaccination (4). Immunological, viral, environmental and host genetic variables affect the progression of HBV infection (5). In particular, the host genetic variables have received increasing attention, as the effects they have may play a role in determining the severity of infection with HBV (6). Extensive research has examined the mechanisms through which genetic variations, such as those in the genes for cytokines, chemokines and human leukocyte antigen, affect vulnerability to herpes simplex virus infection. A new component in the pathophysiology of HBV infection is cytokines, which are key regulators of hepatocyte function and the immune response (7). There has been growing recognition that polymorphisms within cytokine genes may play a role in the observed variation in illness outcomes (8). It has been suggested that the inflammatory cytokine, interleukin-6 (IL)-6 plays a major role in the onset and advancement of liver disorders (9). Studies have shown that blood IL-6 levels are markedly higher in patients with toxic hepatitis, alcoholic or non-alcoholic cirrhosis, and in non-control subjects compared to healthy subjects (10). Another finding is that CD4+ T-cells from individuals with acute-on-chronic HBV liver failure produce more IL-6(11). Monitoring the plasma levels of IL-6 in patients with HCC could help to estimate the disease severity, as an elevated plasma level of IL-6 indicates a poor prognosis (12). Through stimulating the STAT3 signaling pathway, IL-6 promotes HCC cell proliferation and migration (13). It has been proven that IL-6 expression-regulating variables are crucial in the aetiology of liver disorders. Inherited variants in the IL-6 gene may modulate its levels of expression, and those with the CC genotype of the rs1800796 polymorphism has a greater amount of IL-6 mRNA compared to people with the CG/GG genotypes (14).
As the present study was a meta-analysis, the exact molecular processes that cause hepatocarcinogenesis in relation to HBV were not uncovered. The unfinished double-stranded DNA molecule known as the HBV genome is divided into four main regions: The precore area (15), the enhancer II (EnhII) region (16), the basal core promoter (BCP) region (17) and the PreS/S region (18). Notably, there is consistent evidence linking mutations in these particular regions, the PreS, EnhII, BCP and precore regions, to the development of HCC (19). Human leukocyte antigen (HLA), cytokines, and chemokines are only a few of the host genes linked to viral clearance or persistence in infection, according to a number of studies. This genetic predisposition has been greatly illuminated by genome-wide association studies (GWAS). In addition, new HCC risk loci on chromosomes 6p21.32 and 21q21.3 have been found by GWAS in patients with chronic HBV infection. Accordingly, the present systematic review and meta-analysis was conducted to determine the association between the IL-6 (rs1800796) gene polymorphism and susceptibility to HBV and HCC.
Data and methods
Search strategy
Experimental case-control studies have been performed to investigate the possible association between the IL-6 polymorphism, rs1800796, and the risk of developing HBV infection and HCC. A systematic literature search that utilized the PubMed, Embase and Google Scholar databases was made. This search was narrowed down to literature published in the English language and was carried out until December 1, 2024. The following search terms were used: rs1800796; hepatocellular carcinoma; liver cancer; polymorphism; SNP; genotype. Gene polymorphism studies on IL-6 in HBV and HCC were conducted in China (14,20,21,22), Malaysia (23), Egypt (24,25) and India (26). Furthermore, relevant articles were located by manually searching the reference lists of the selected articles and reviews.
Inclusion and exclusion criteria
Studies that focused on the IL-6 gene polymorphisms (rs1800796) and the risk of developing HBV and HCC in individuals as a case-control methodology was included. The following criteria were included in the meta-analysis: Availability of the full-text article for the complete information to determine an odds ratio (OR) with a 95% confidence interval (CI), the investigation reports containing the genotype frequencies and the research publication on diverse populations.
Studies were not included unless they satisfied the following criteria: Studies pertaining to IL-6 genetic variations and their link to susceptibility to HBV and HCC; studies using animal models; studies providing genotypic and allelic frequency data; and articles including abstracts and reviews not clinically in character.
Data extraction
Following the inclusion criteria, data were collected. Discussions with co-authors helped to resolve any differences. Published research was examined to identify the genotypes and allele frequencies in the case and control groups. Information, such as first author's name, Hardy-Weinberg Equilibrium (HWE) score, PubMed ID, language, year of publication, ethnicity, sample size and study design was recorded. The prototype test was conducted for a data extraction to ensure the uniformity and the screening flexibility.
Statistical analysis
The association between polymorphisms in the IL-6 genes and the risk of HBV/HCC were investigated in a number of statistical methods. ORs and 95% CI values were estimated to project the connection. Statistical significance was assessed using a threshold of P<0.05. Different genetic models namely over-dominant, dominant, recessive and allelic were analyzed in the present meta-analysis. Heterogeneity among studies was evaluated by I2 based Q-statistics. An I2 <25% was usually viewed as low heterogeneity, between 25 and 50% as moderate, and >50% as high heterogeneity. The I2 value is independent of the number of studies included in the meta-analysis, but it actually quantifies the effect of heterogeneity. Therefore, pooled ORs were calculated for the three models using a random effects model due to the high degree of heterogeneity. Two investigations were found to be dissimilar statistically. Using the Q statistic and I2, homogeneity across studies was studied; summary odds ratios were looked at using a Z-test (P<0.05). A sensitivity analysis was conducted to probe the effects of excluding certain trials, particularly those in which the controls did not follow HWE. Egger's regression method was also used in search of publication bias. Statistical analysis was carried using MetaGenyo software (https://metagenyo.genyo.es/).
Assessment of quality of studies using risk bias and protein-protein interaction (PPI)
A meta-analysis is a careful assessment of both the quality of the technique and the potential biases in studies. The present study used the Cochrane Risk of Bias tool (ROB2) to evaluate the possible bias in the included studies. This assessment labelled the studies as either ‘low risk’, ‘some concern’, or ‘high risk’ of bias. The STRING (v11.0) online database can predict functional proteins and PPIs related to identified IL-6 associated polymorphisms, with a confidence score of at least 0.4.
Results
Study identification and selection
A total of eight distinct studies (14,20-26) were considered following the removal of duplication from 144 articles obtained from the PubMed, Embase and Google Scholar databases. A total of 4,767 publications were omitted in the screening procedure that relied on abstracts and titles. Review articles, studies on other genes, studies examining the association between sMICA expression and HCC, and studies on other illnesses were all ineligible. Following a careful examination of the 144 articles, 20 articles were complete texts; 12 of these publications were excluded, due to issues such being corrigenda, not having data that could be extracted, not having any control groups, having data that were duplicated, or having information about viral infections that was not clear. Finally, the meta-analysis comprised of eight articles that satisfied the criteria (Fig. 1).
General characteristics of the included studies
There were 3,630 control subjects, 1,960 patients with HBV, and 877 patients with HCC included in the eight articles that comprised the meta-analysis. Individuals infected with HBV were examined in the included studies, and they were of both Asian and Caucasian ethnicities. Furthermore, the HWE was supported by the SNP data from the control groups of all eight studies. A comprehensive summary of the listed studies and the basic features is presented in Tables I and II.
Table IDistribution of the IL-6 rs1800796 allele and genotype among HBV cases and controls, and Hardy-Weinberg equilibrium score. |
Table IIDistribution of the IL-6 rs1800796 allele and genotype among HCC cancer cases and controls, and the Hardy-Weinberg equilibrium score. |
Meta-analysis of the association between IL-6 (rs1800796) polymorphism and the risk of developing HBV
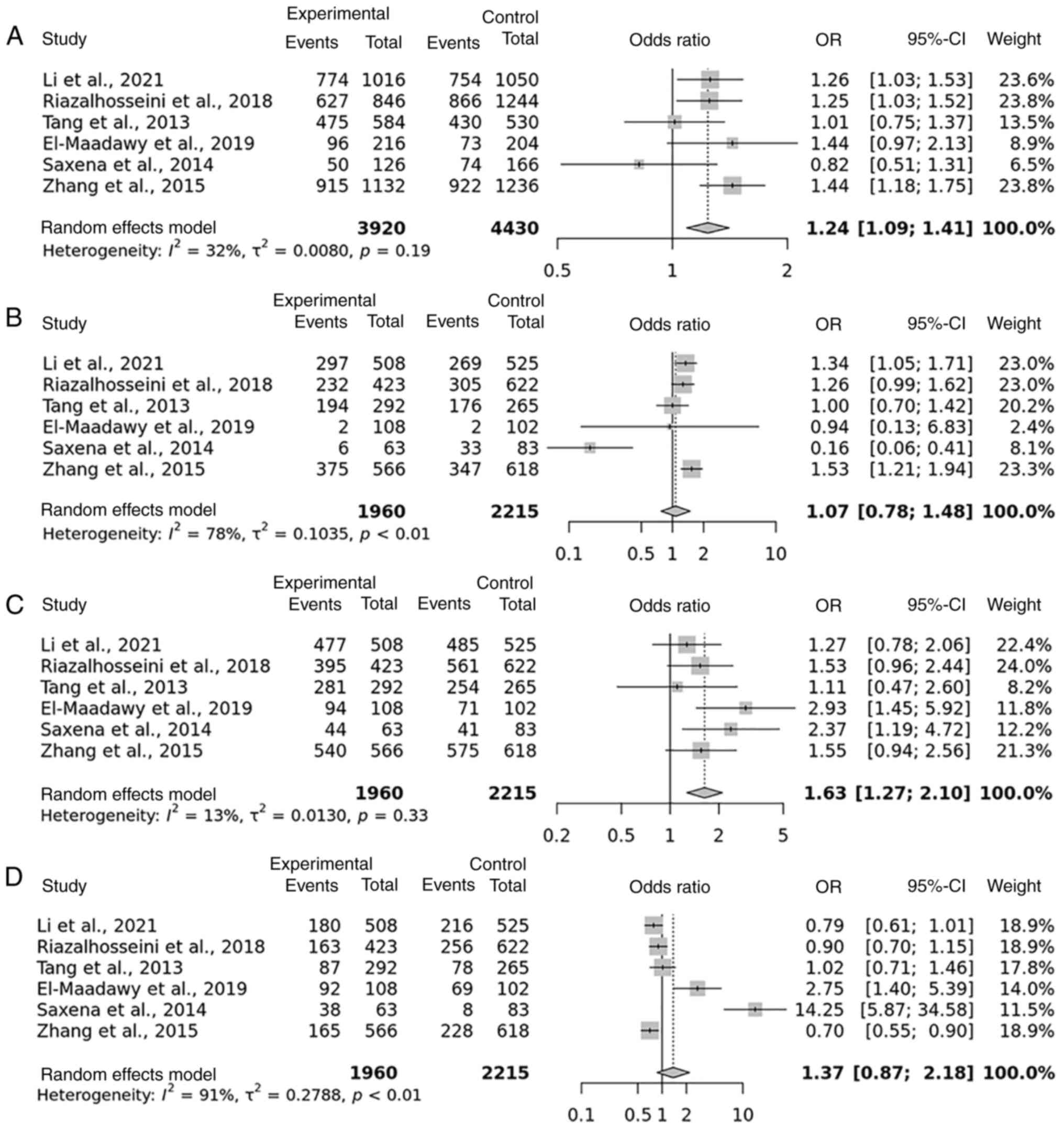
A summary of the findings of the meta-analysis regarding the potential link between the rs1800796 polymorphism and risk of developing HBV is presented in Table I. HBV was included in all five studies that examined the rs1800796 polymorphism; a total of 1,960 cases and 2,215 controls were included. In the allelic model, a random-effects meta-analysis of the whole population revealed a significant association between the rs1800796 polymorphism and an elevated risk of developing HBV (P=0.01). The results for the dominant CG genotype of rs1800796 in patients with HBV were similar with a level of significance of P=0.01. For both the over-dominant and recessive models, the results indicated a insignificant value (P=0.17 and P=0.67, respectively) (Table SI). These data suggest that there is an association between the CG genotype of the allelic and dominant models of the rs1800796 polymorphism and the risk of developing HBV in Asian and Caucasian populations (Fig. 2). A summary of the findings of the meta-analysis for the rs1800796 polymorphism in the Asian and Caucasian populations as regards susceptibility to HBV is presented in Table I.
Meta-analysis of the association between the IL-6 (rs1800796) polymorphism and the risk of developing HCC
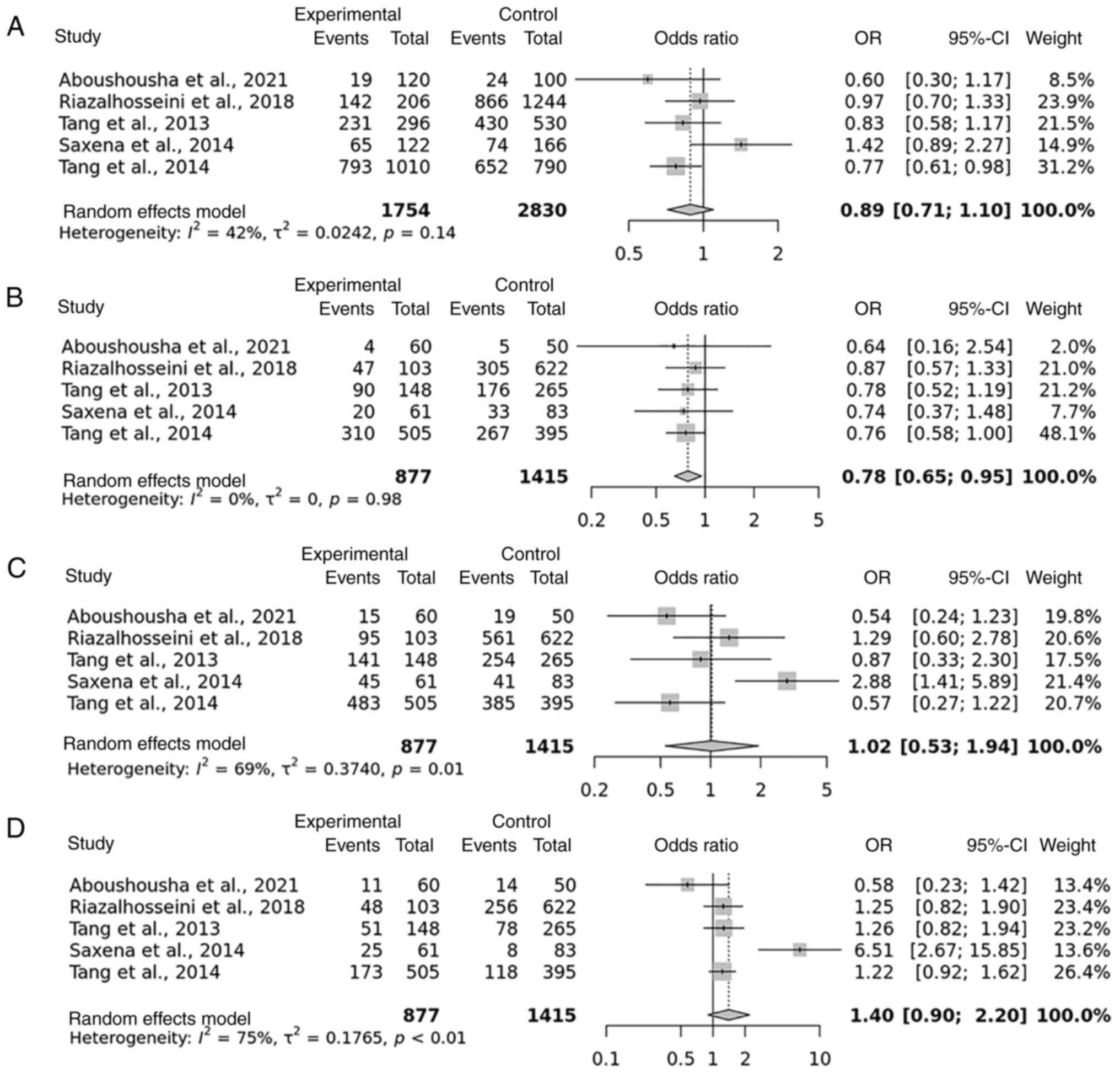
The results of the meta-analyses of the potential link between the rs1800796 polymorphism and the risk of developing HCC are summarized in Table II. The rs1800796 polymorphism did not exhibit a significant association with HCC in either the allelic, dominant and over- dominant models (P=0.27, P=0.95 and P=0.13, respectively) in the overall sample of 877 cases and 1,415 controls. However, in the recessive genetic model, the rs1800796 polymorphism was linked to an increased risk of susceptibility to HCC (P=0.01) (Table SII) According to these findings, there was an increased risk of HCC associated with the CG genotype of the recessive genetic model in rs1800796 in Asian and Caucasian populations (Fig. 3).
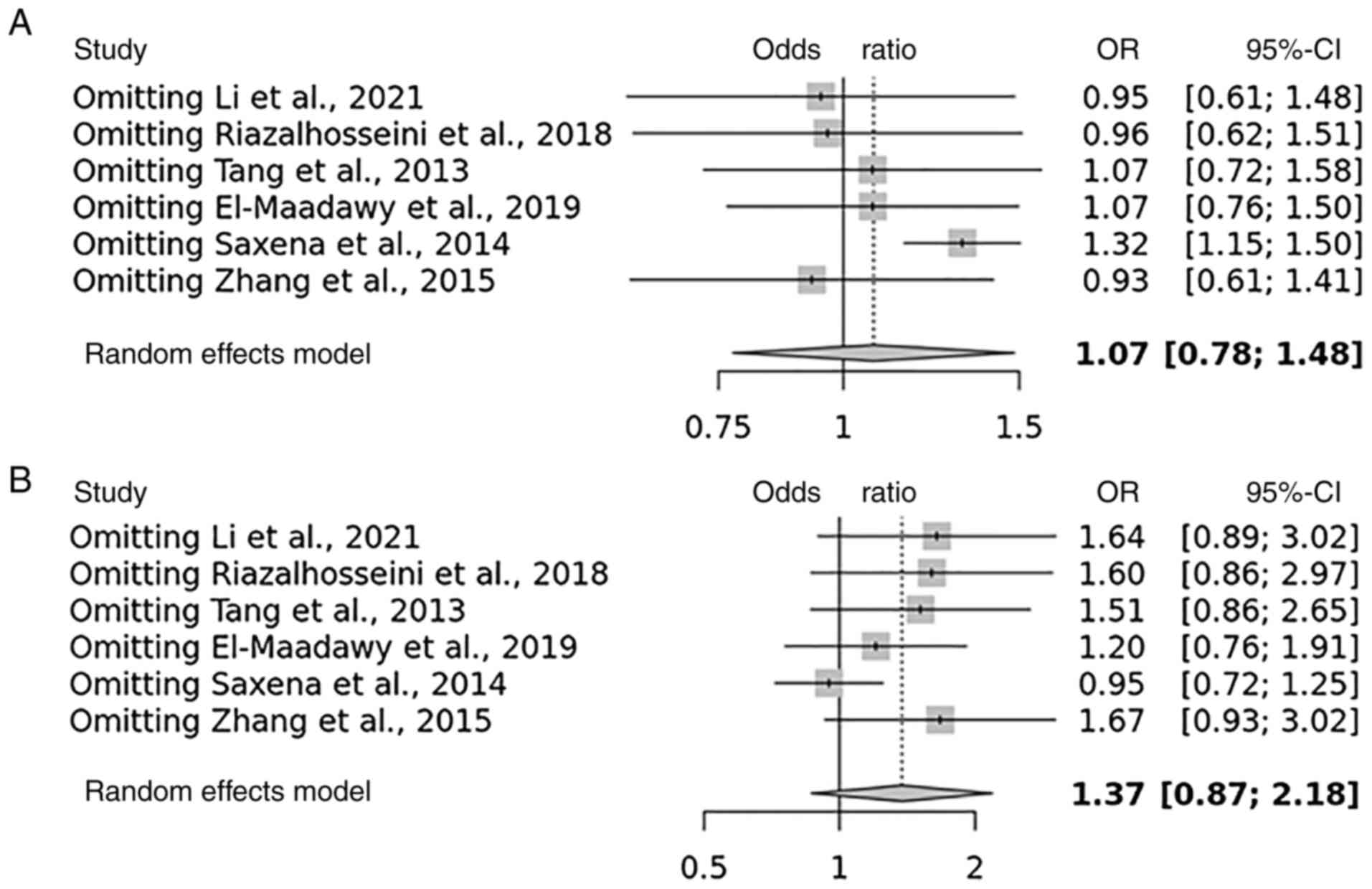
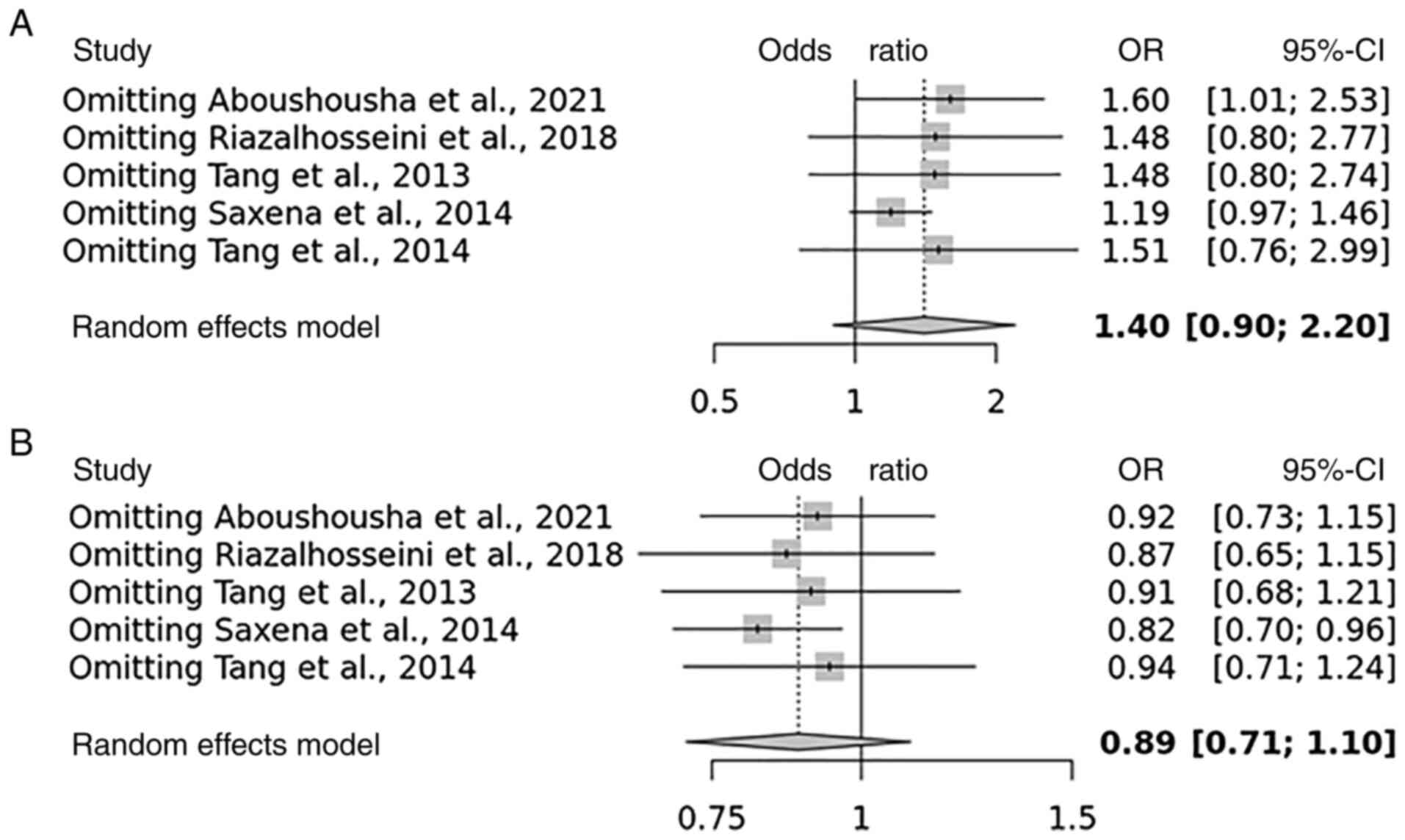
Sensitivity analysis, publication bias and risk of bias
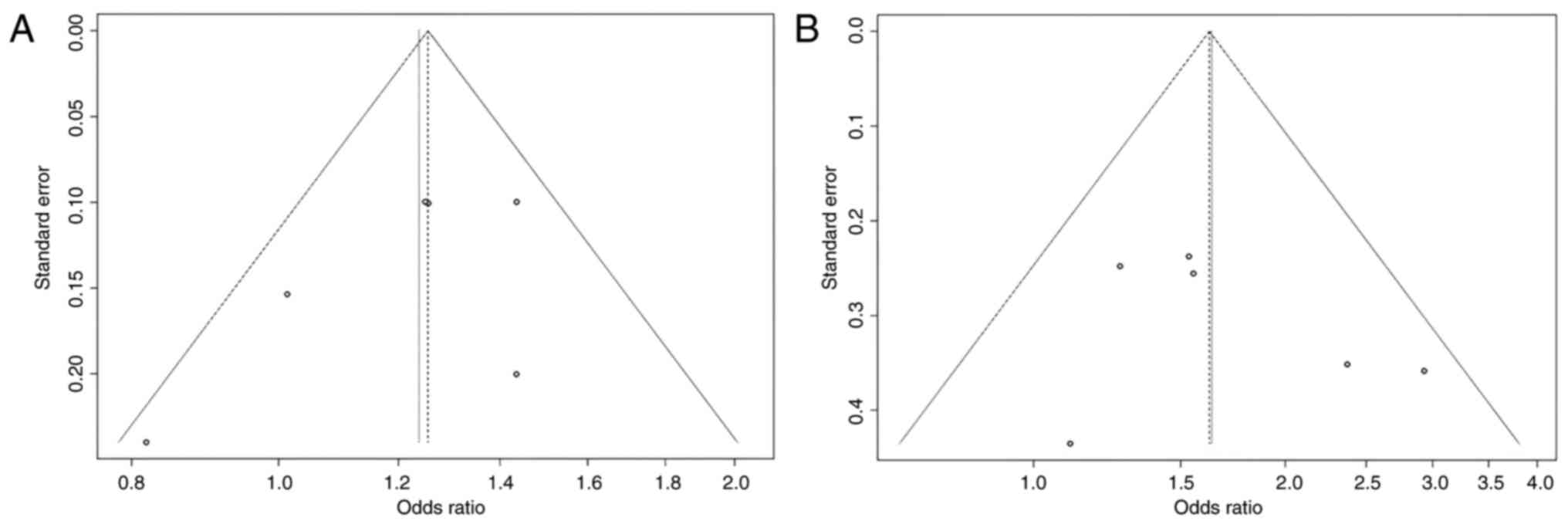
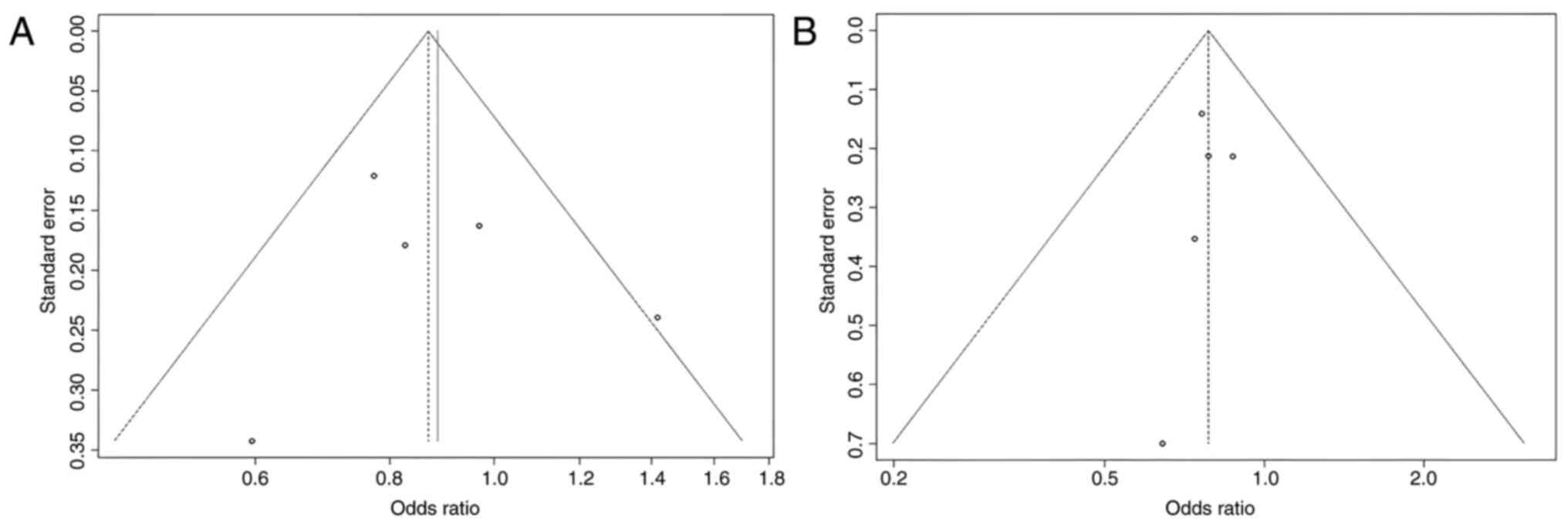
Some studies had contradictory results, which may explain why the total results were so varied, according to the HWE. The present study identified that four studies are with unbalanced case-control ratios as possible causes of heterogeneity. A sensitivity analysis was carried out to determine the influence of possible differences in intervention effects and heterogeneity (Figs. 4 and 5). Studies that did not follow the intervention standards or did not follow HWE were not included in this analysis. Of note, the final P-value was unaltered even after excluding individual studies. To assess the possibility of publication bias, a funnel plot was used (Figs. 6 and 7).
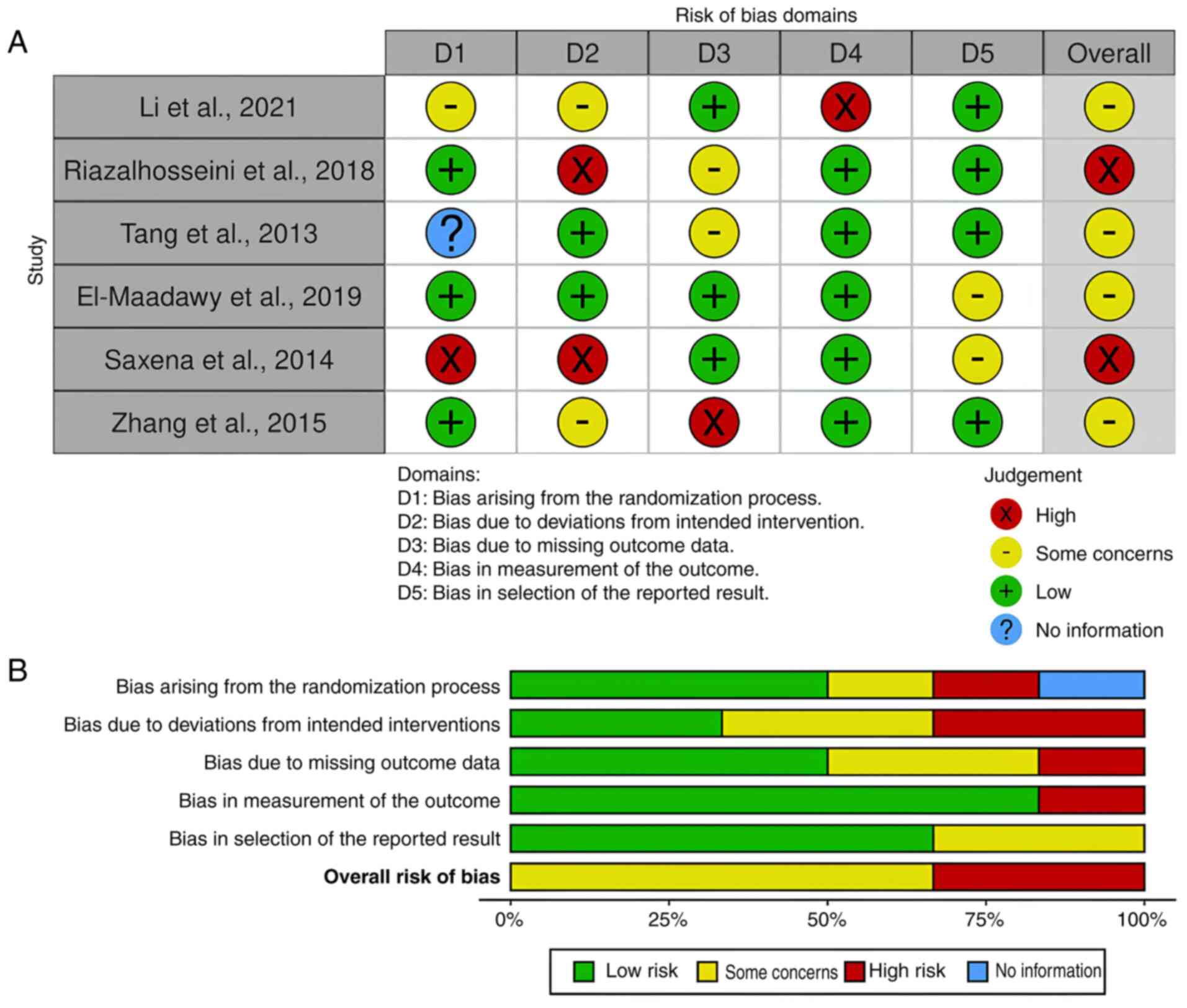
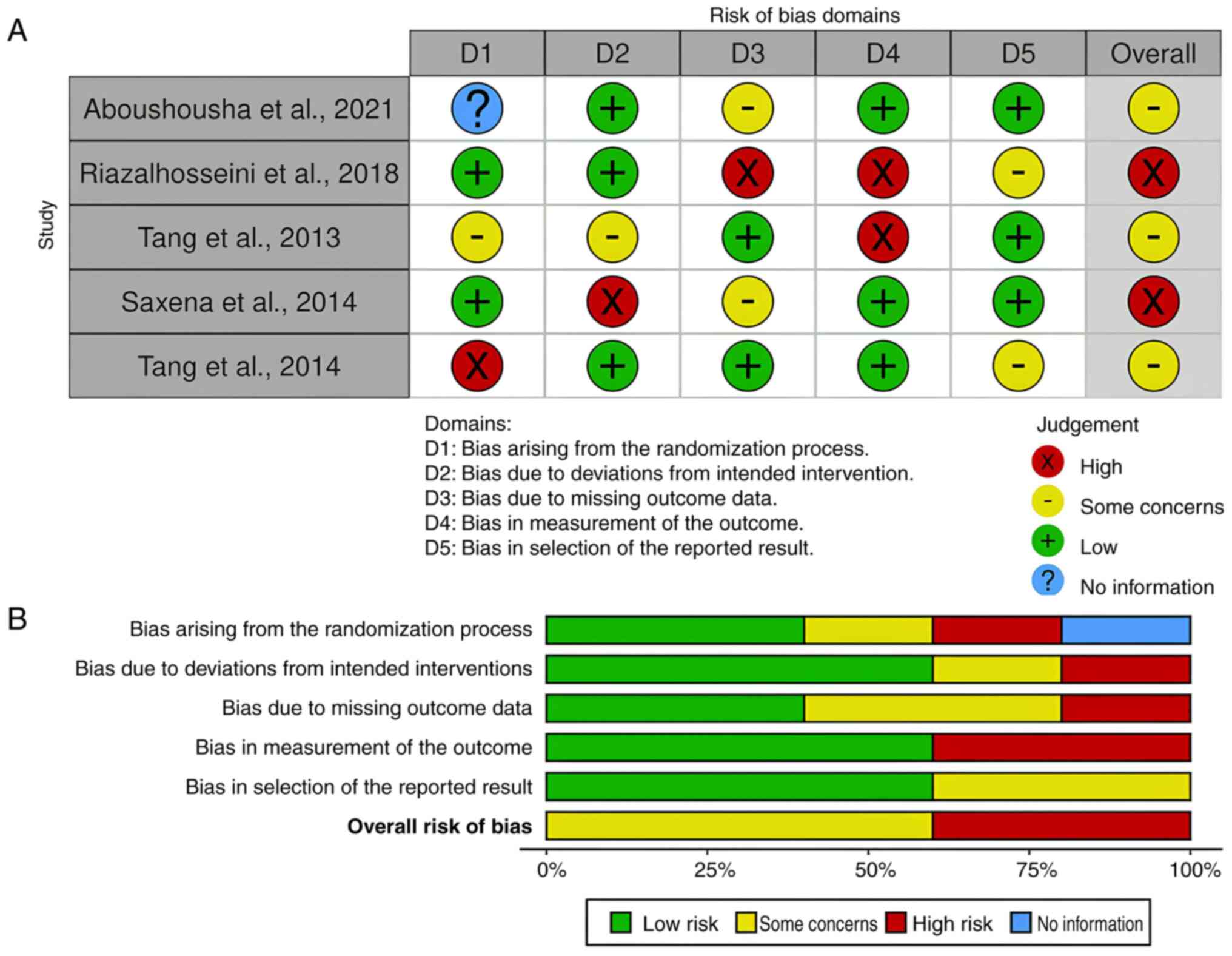
The Cochrane Risk of Bias Tool 2 was used to evaluate the methodological quality of every included study comprehensively. Each study topic has a row and columns for the many bias categories. The degree of risk associated with each type of bias in each type of study is illustrated in Figs. 8 and 9 with a color-coded system. Red denotes a great risk, yellow a moderate risk and green a modest risk. The majority of studies revealed some concerns as regards risk of bias, which is in line with well-documented techniques meant to lower the possibility of bias or systematic errors.
Power analysis and construction of the PPI network
A power analysis was performed to ascertain the significance level of each study for the selected SNPs. The sample size of the selected literature was adequate to satisfy the significant level requirements, which include an α error probability of 0.05. The computation of the power results is presented in Table III. The power analysis plot revealed the findings of the power analysis for a two-tailed hypothesis test. Using power analysis, one can assess the likelihood of finding an influence of a certain magnitude under specific conditions, that is, a given sample size, effect size and significance threshold.
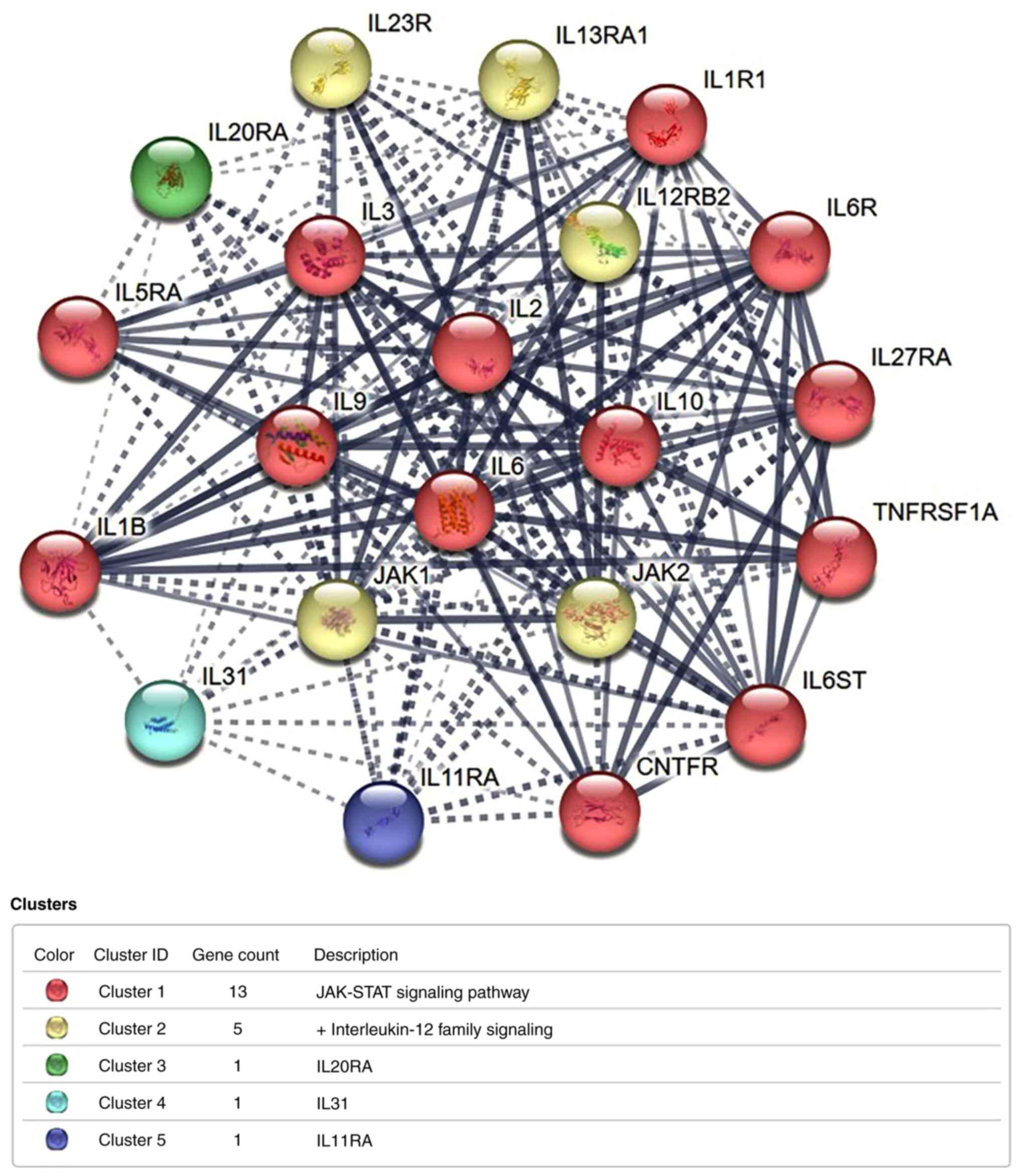
To find the action of genes as hubs for the polymorphic proteins linked to HBV and HCC, the STRING approach was used to map and investigate these (Figs. 10 and 11). The IL-06 protein network boasts 21 nodes and 170 edges with a PPI enrichment P-value of P<1.0e-16 and a clustering coefficient of 0.884. The average of node degrees was 16.2. The protein network had more interactions with its constituents than was expected, while it was compared to the random choice of proteins of similar degree and size. This higher enrichment implies that these proteins work together and they are physiologically linked.
Discussion
The present meta-analysis aimed to determine the association of a IL-6 SNP with the onset of HBV and HCC. The results revealed that the IL-6 rs1800796 SNP is the potential hereditary variable affecting the likelihood of developing liver disorders. Liver illnesses, including HBV and HCC, are more common among Asian and Caucasian communities where individuals with the CG or the GG + GC, or GG, GC genotypes of rs1800796 are more likely to develop liver illnesses. On the other hand, the G allele, GG, or GC of genotypes may provide some protection against the onset of liver disorders, including HBV and HCC, particularly in Asian populations.
The IL-6 gene promoter is situated 174 base pairs upstream of the rs1800795 polymorphism (27). Researchers have demonstrated that lowering the transcription rate of the gene, which in turn lowers IL-6 production, can be achieved by changing this codon from G to C (28). It has been shown that hepatitis B core antigen transfection has the potential to enhance IL-6 synthesis and release in hepatocytes by activating signaling pathways, such as extracellular signal-related kinase, nuclear factor-κB and p38 mitogen-activated protein kinase (29). Hepatitis B core antigen transfection enhances IL-6 expression and secretion in hepatocytes, according to previous research (30). When the STAT3 pathway is activated, for example, by IL-6 produced by HBV, the transcription and translation of genes encoding vascular endothelial growth factor and angiogenin can begin (31). Patients with alcoholic liver damage and non-alcoholic steatohepatitis have increased levels of inflammatory and oxidative stress markers, suggesting that this activation may cause these conditions (32). Serum IL-6 levels and the IL-6 gene polymorphism (rs1800796) have been studied in relation to the development of chronic HBV infection; there is a clear association between higher blood IL-6 levels and the course of illness, particularly in the late stages of liver disease (33). IL-6 expression appears to be more critical than genetic variations in the development of chronic HBV infection.
Chronic HBV and its consequences for the liver have been shown to be more or less likely to occur in individuals with certain IL-6 gene variations (34). IL-6 gene polymorphisms can affect both the susceptibility to and the development of HBV infection. A previous study examined the possibility that certain variations in the IL-6 gene would increase the likelihood that an individual would become infected with HBV. Some IL-6 gene variations are associated with a decreased likelihood of acquiring chronic HBV infection. IL-6 gene polymorphisms could affect the course of illness and the host immunological response to herpes simplex virus infection (35).
GWAS have demonstrated that SNPs in the IL-6 gene significantly increase the risk of developing several malignancies. A previous meta-analysis of 118 GWAS including 50,053 cases and 65,204 control samples revealed substantial evidence relating the IL-6 gene rs1800796 to an increased risk of developing cancer (36). The rs1800795 polymorphism was linked to higher risk of developing prostate, liver and cervical cancer. Subgroup analysis based on ethnicity revealed that the genomic regions rs1800795, rs1800796 and rs1800797 were substantially linked, respectively, to an increased risk of developing cancer in African, Asian and Pacific Islander populations (36).
Two-stage meta-GWAS of circulating blood IL-6 levels revealed independence between the HLA-DRB1/DRB5 rs660895 locus on chromosome 6p21 and the IL1F10/IL1RN rs6734 locus on chromosome 2q14. Furthermore, the investigation was repeated for conformation of the IL6R rs4537545 gene on 1q21 of chromosome. These recently identified immunological and inflammatory routes may influence the pathobiology of IL-6(37).
Studies have linked several IL6R gene variants to the risk of developing inflammatory, autoimmune and cardiovascular diseases, and higher levels of inflammatory biomarker. Of note, 10 distinct variants in this gene have clearly been linked to a concentration of four different phenotypes, four different inflammatory diseases, and an elevated risk of five different cardiovascular diseases; five SNPs have also been shown to be related to one another: rs2228145, rs7518199, rs4537545, rs7529229 and rs4129267(38). Liu et al (39) stated that IL-6 was not associated with HCC, whereas the present study revealed the association of IL-6 with HCC in a recessive genetic model. These findings align with prior meta-analyses, indicating a possible elevated risk of developing HCC in individuals carrying the G allele of the IL-6-174 G/C polymorphism, but not the IL-6-572 G/C. A significant gap exists in the understanding of the effects of the IL-6 gene polymorphism on HCC in the context of HBV infection. The present study addresses this gap through a dedicated case-control analysis. Numerous factors, including the biological structure of the IL-6 polymorphism, the population heterogeneity and sample size distribution can affect the different significance among the allelic, recessive, dominant and over-dominant models. Different statistical power results from the fact that each model investigates different inheritance hypotheses. For certain cases, the recessive model could be relevant; however, the dominant model is not as it detects an influence by homozygosity of the minor allele. This may be the result of different genetic associations or disequilibrium linkage patterns between the subpopulations that were examined in the meta-analysis generating model-dependent effects. Likewise, the over-dominant model indicates a heterozygote advantage, whereas the recessive model indicates that homozygous individuals drive the effect. These findings imply that complex genetic or regulatory mechanisms affect IL-6 expression; thus, future studies are warranted to investigate these systems in further detail.
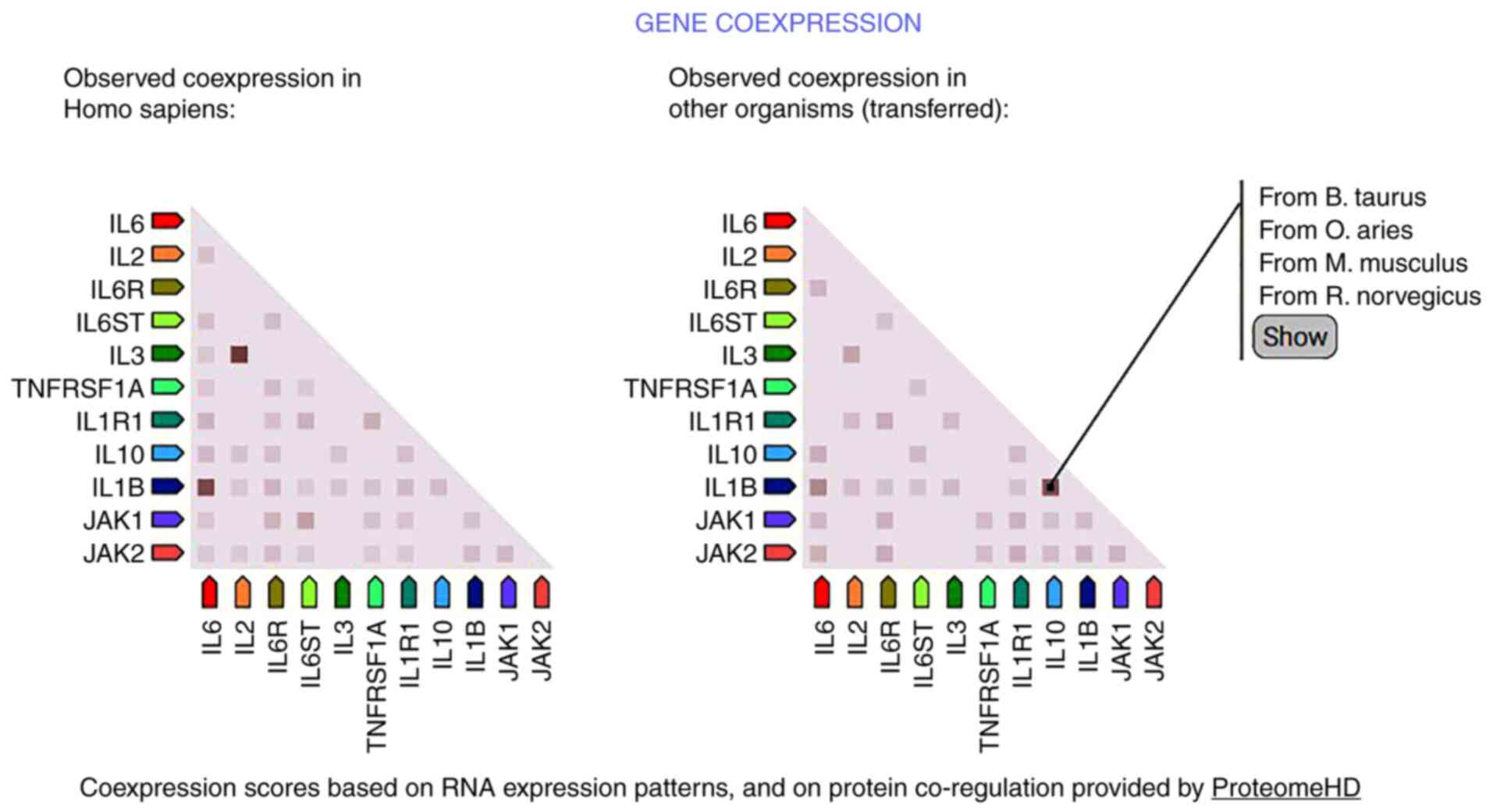
The present study investigated the association between the rs1800796 polymorphism and susceptibility to HBV and HCC. HBV susceptibility was not associated with any of the genetic variables in the IL-6 (rs1800796) gene polymorphism. Data extraction and analysis were conducted in accordance with rigorous techniques. A power analysis was executed to assure the large enough samples to obtain the significant findings (Table III). The results on HCC using the IL-6 (rs1800796) gene and SNPs revealed that only one distinct variant significantly increased the risk of developing the disease. The present meta-analysis included studies that involved both cases and controls of HBV and HCC. This analysis utilized the different raw data and produced results that differed from the results of the research articles (20,24). Individuals with the CG genotype at rs1800796 had an increased risk of developing HBV and HCC, particularly in Asian populations. This indicates that environmental variables, such as viral infection and geographic location, play a role in determining susceptibility to certain illnesses, in addition to individual genetic factors. Factors including viral genotype, sex and comorbidities may explain the heterogeneity retained following subgroup analysis. The quantitative Egger's test analysis and qualitative Begg's test plot were used to evaluate publication bias. The combined findings are reliable, as neither test revealed a substantial publication bias. PPI network analysis of polymorphic IL-6-associated proteins made use of the STRING database. The developed IL-6 gene network consisted of 21 nodes and 170 edges overall. Co-expression research revealed that IL-6 exhibited direct interactions within the network, and the IL-1 gene, mediated via other genes, had the connection.
Nevertheless, there some limitations to the present study. One potential issue is that publishing and cultural biases may have been created due to the inclusion of solely English-language articles. As a second point, the results may not be applicable to individuals of other ethnicities, since the majority of genotyping data were derived from Chinese populations.
In conclusion, the present systematic review and meta-analysis on the association between the IL-6 rs1800796 genetic variant and the severity of HBV and HCC revealed contradicting data. Although early statistical data were connected with variance to disease risk, a comprehensive meta-analysis revealed no significant association between HBV severity across various genetic variants. Still, it was revealed that HCC was associated with the IL-6 rs1800796 gene polymorphism in a recessive genetic model. A number of genes scattered around the genome define a complex trait that renders a person susceptible to HBV and HCC. The degree of the impact of these genes is dependent on several factors, including the study population, case-control selection criteria and sample size. Studies with a few samples may lead to weak findings. Only large-scale, randomized studies covering a broad spectrum of individuals can provide reliable evidence associating genetic variations to HBV/HCC. Precision medicine offers possibilities for tailored prevention and early diagnosis, especially for therapies aiming targeting IL-6.
Supplementary Material
HBV genetic models.
HCC genetic models.
Acknowledgements
The authors would like to thank the management of Chettinad Academy of Research and Education (Deemed to be University), Kelambakkam, India, for providing the facilities to perform this study.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
BSA conducted the literature search, collected data, developed the study methodology, contributed to the writing of the manuscript, and created the tables and figures. KM was involved in the writing of the original draft of the manuscript, in data validation, and in data curation. AR and YM has involved in the study design, data analysis, preparation of the manuscript, interpretation of the results, editing assistance, study supervision, and final drafting. GKS conducted the investigations, provided editing assistance, supervised the study, and was also involved in the conceptualization of the study. The authors confirm that all raw data presented in this article authentic and accurately represent the finding of study. BS and GKS confirm the authenticity of all the raw data. All authors have thoroughly reviewed and have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Madihi S, Syed H, Lazar F, Zyad A and Benani A: A systematic review of the current hepatitis B viral infection and hepatocellular carcinoma situation in Mediterranean countries. Biomed Res Int. 2020(7027169)2020.PubMed/NCBI View Article : Google Scholar | |
|
Soriano V, Moreno-Torres V, Treviño A, de Jesús F, Corral O and de Mendoza C: Prospects for controlling hepatitis B globally. Pathogens. 13(291)2024.PubMed/NCBI View Article : Google Scholar | |
|
Sun H, Chang L, Yan Y and Wang L: Hepatitis B virus pre-S region: Clinical implications and applications. Rev Med Virol. 31(e2201)2021.PubMed/NCBI View Article : Google Scholar | |
|
Lim YS, Kim WR, Dieterich D, Kao JH, Flaherty JF, Yee LJ, Roberts LR, Razavi H and Kennedy PT: Evidence for benefits of early treatment initiation for chronic hepatitis B. Viruses. 15(997)2023.PubMed/NCBI View Article : Google Scholar | |
|
Ma H, Yan QZ, Ma JR, Li DF and Yang JL: Overview of the immunological mechanisms in hepatitis B virus reactivation: Implications for disease progression and management strategies. World J Gastroenterol. 30:1295–1312. 2024.PubMed/NCBI View Article : Google Scholar | |
|
Pollicino T and Caminiti G: HBV-integration studies in the clinic: Role in the natural history of infection. Viruses. 13(368)2021.PubMed/NCBI View Article : Google Scholar | |
|
Iannacone M and Guidotti LG: Immunobiology and pathogenesis of hepatitis B virus infection. Nat Rev Immunol. 22:19–32. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Xu J, Zhan Q, Fan Y, Yu Y and Zeng Z: Human genetic susceptibility to hepatitis B virus infection. Infect Genet Evol. 87(104663)2021.PubMed/NCBI View Article : Google Scholar | |
|
Aliyu M, Zohora FT, Anka AU, Ali K, Maleknia S, Saffarioun M and Azizi G: Interleukin-6 cytokine: An overview of the immune regulation, immune dysregulation, and therapeutic approach. Int Immunopharmacol. 111(109130)2022.PubMed/NCBI View Article : Google Scholar | |
|
Neuman MG, Mueller J and Mueller S: Non-invasive biomarkers of liver inflammation and cell death in response to alcohol detoxification. Front Physiol. 12(678118)2021.PubMed/NCBI View Article : Google Scholar | |
|
Zhou C, Zhang N, He TT, Wang Y, Wang LF, Sun YQ, Jing J, Zhang JJ, Fu SN, Wang X, et al: High levels of serum interleukin-6 increase mortality of hepatitis B virus-associated acute-on-chronic liver failure. World J Gastroenterol. 26:4479–4488. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Rico Montanari N, Anugwom CM, Boonstra A and Debes JD: The role of cytokines in the different stages of hepatocellular carcinoma. Cancers. 13(4876)2021.PubMed/NCBI View Article : Google Scholar | |
|
Xu J, Lin H, Wu G, Zhu M and Li M: IL-6/STAT3 is a promising therapeutic target for hepatocellular carcinoma. Front Oncol. 11(760971)2021.PubMed/NCBI View Article : Google Scholar | |
|
Li M, Zhuo Y, Xu Y, Chen H, Cheng Z and Zhou L: Genetic association of interleukin-6 polymorphism (rs1800796) with chronic hepatitis B virus infection in Chinese Han population. Viral Immunol. 34:267–272. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Li W, Wang S, Jin Y, Mu X, Guo Z, Qiao S, Jiang S, Liu Q and Cui X: The role of the hepatitis B virus genome and its integration in the hepatocellular carcinoma. Front Microbiol. 15(1469016)2024.PubMed/NCBI View Article : Google Scholar | |
|
Qu B and Brown RJ: Strategies to inhibit hepatitis B virus at the transcript level. Viruses. 13(1327)2021.PubMed/NCBI View Article : Google Scholar | |
|
Farshadpour F, Taherkhani R and Saberi F: Molecular evaluation of hepatitis B virus infection and predominant mutations of pre-core, basal core promoter and S regions in an Iranian population with type 2 diabetes mellitus: A case-control study. BMC Infect Dis. 22(553)2022.PubMed/NCBI View Article : Google Scholar | |
|
Xie C and Lu D: Evolution and diversity of the hepatitis B virus genome: Clinical implications. Virology. 598(110197)2024.PubMed/NCBI View Article : Google Scholar | |
|
Zhao Y, Chen K, Yang H, Zhang F, Ding L, Liu Y, Zhang L, Zhang Y, Wang H and Deng Y: HLA-DR genetic polymorphisms and hepatitis B virus mutations affect the risk of hepatocellular carcinoma in Han Chinese population. Virol J. 20(283)2023.PubMed/NCBI View Article : Google Scholar | |
|
Tang S, Liu Z, Zhang Y, He Y, Pan D, Liu Y, Liu Q, Zhang Z and Yuan Y: Rather than Rs1800796 polymorphism, expression of interleukin-6 is associated with disease progression of chronic HBV infection in a Chinese Han population. Dis Markers. 35:799–805. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Tang S, Yuan Y, He Y, Pan D, Zhang Y, Liu Y, Liu Q, Zhang Z and Liu Z: Genetic polymorphism of interleukin-6 influences susceptibility to HBV-related hepatocellular carcinoma in a male Chinese Han population. Hum Immunol. 75:297–301. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Zhang G, Wang W, Li S, Yang H, Zhang M, Zhang P, Wen Y, Wu A, Yang L, Zhou B and Chen X: IL6 gene allele-specific C/EBPα-binding activity affects the development of HBV infection through modulation of Th17/Treg balance. Genes Immun. 16:528–535. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Riazalhosseini B, Mohamed Z, Apalasamy YD, Shafie NS and Mohamed R: Interleukin-6 gene variants are associated with reduced risk of chronicity in hepatitis B virus infection in a Malaysian population. Biomed Rep. 9:213–220. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Aboushousha T, Emad M, Rizk G, Ragab K, Hammam O, Fouad R and Helal NS: IL-4, IL-17 and CD163 immunoexpression and IL-6 gene polymorphism in chronic hepatitis C patients and associated hepatocellular carcinoma. Asian Pac J Cancer Prev. 22:1105–1113. 2021.PubMed/NCBI View Article : Google Scholar | |
|
El-Maadawy EA, Talaat RM, Ahmed MM and El-Shenawy SZ: Interleukin-6 promotor gene polymorphisms and susceptibility to chronic hepatitis B virus in Egyptians. Hum Immunol. 80:208–214. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Saxena R, Chawla YK, Verma I and Kaur J: IL-6 (-572/-597) polymorphism and expression in HBV disease chronicity in an Indian population. Am J Hum Biol. 26:549–555. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Ren H, Guo Z, Qin WJ and Yang ZL: Association of interleukin-6 genetic polymorphisms (rs1800795,-174C> G and rs1800796,-572G> C) with risk of essential hypertension in the Chinese population. Cureus. 15(e46334)2023.PubMed/NCBI View Article : Google Scholar | |
|
Almolakab ZM, El-Nesr KA, Mohamad Hassanin EH, Elkaffas R and Nabil A: Gene polymorphisms of Interleukin 6 (-174 G/C) and transforming growth factor β-1 (+915 G/C) in ovarian cancer patients. Beni-Suef Univ J Basic Appl Sci. 11(30)2022. | |
|
Zheng S, Qi W, Xue T, Zao X, Xie J, Zhang P, Li X, Ye Y and Liu A: Chinese medicine in the treatment of chronic hepatitis B: The mechanisms of signal pathway regulation. Heliyon. 10(e39176)2024.PubMed/NCBI View Article : Google Scholar | |
|
Kong F, Tao Y, Yuan D, Zhang N, Li Q, Yu T, Yang X, Kong D, Ding X, Liu X, et al: Hepatitis B virus core protein mediates the upregulation of C5α receptor 1 via NF-κB pathway to facilitate the growth and migration of hepatoma cells. Cancer Res Treat. 53:506–527. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Tavakoli Pirzaman A, Alishah A, Babajani B, Ebrahimi P, Sheikhi SA, Moosaei F, Salarfar A, Doostmohamadian S and Kazemi S: The role of microRNAs in hepatocellular cancer: A narrative review focused on tumor microenvironment and drug resistance. Technol Cancer Res Treat. 23(15330338241239188)2024.PubMed/NCBI View Article : Google Scholar | |
|
Bovind AP, Marciano F, Mandato C, Siano MA, Savoia M and Vajro P: Oxidative stress in non-alcoholic fatty liver disease. An updated mini review. Front Med (Lausanne). 8(595371)2021.PubMed/NCBI View Article : Google Scholar | |
|
Zhong S, Zhang T, Tang L and Li Y: Cytokines and chemokines in HBV infection. Front Mol Biosci. 8(805625)2021.PubMed/NCBI View Article : Google Scholar | |
|
Elbrolosy AM, Elabd NS, ElGedawy GA, Abozeid M, Abdelkreem M, Montaser B, Eed EM and Elhamouly MS: Toll-like receptor 2 polymorphism and IL-6 profile in relation to disease progression in chronic HBV infection: A case control study in Egyptian patients. Clin Exp Med. 23:117–129. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Abd El-Baky RM, Hetta HF, Koneru G, Ammar M, Shafik EA, Mohareb DA, Abbas El-Masry M, Ramadan HK, Abu Rahma MZ, Fawzy MA and Fathy M: Impact of interleukin IL-6 rs-1474347 and IL-10 rs-1800896 genetic polymorphisms on the susceptibility of HCV-infected Egyptian patients to hepatocellular carcinoma. Immunol Res. 68:118–125. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Harun-Or-Roshid M, Ali MB, Jesmin and Mollah MN: Statistical meta-analysis to investigate the association between the Interleukin-6 (IL-6) gene polymorphisms and cancer risk. PLoS One. 16(e0247055)2021.PubMed/NCBI View Article : Google Scholar | |
|
Ahluwalia TS, Prins BP, Abdollahi M, Armstrong NJ, Aslibekyan S, Bain L, Jefferis B, Baumert J, Beekman M, Ben-Shlomo Y and Bis JC: . Genome-wide association study of circulating interleukin 6 levels identifies novel loci. Hum Mol Genet. 30:393–409. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Zhang M, Bai Y, Wang Y, Cui H, Tang M, Wang L, Wang X and Gu D: Cumulative evidence for associations between genetic variants in interleukin 6 receptor gene and human diseases and phenotypes. Front Immunol. 13(860703)2022.PubMed/NCBI View Article : Google Scholar | |
|
Liu Y, Gao SJ, Du BX and Wang JJ: Association of IL-6 polymorphisms with hepatocellular carcinoma risk: Evidences from a meta-analysis. Tumour Biol. 35:3551–3561. 2014.PubMed/NCBI View Article : Google Scholar |