Comparative analysis of vaginitis and endometrial cancer microbiomes using next‑generation sequencing
- Authors:
- Published online on: May 30, 2025 https://doi.org/10.3892/wasj.2025.357
- Article Number: 69
-
Copyright : © Alsaadi et al. This is an open access article distributed under the terms of Creative Commons Attribution License [CC BY 4.0].
Abstract
Introduction
The female reproductive system comprises internal and external organs essential for menstruation and procreation. This organ system is responsible for producing gametes (termed eggs or ova), regulating sex hormones, and maintaining fertilized eggs as they develop into mature fetuses ready for delivery (1).
The vaginal microbiota is a complex group of bacteria that populate the vaginal environment and are vital to maintaining the reproductive health of women. The vaginal microbiota is less diverse than that in other regions of the body and is mostly composed of Lactobacillus species, which produce antimicrobial compounds, including lactic acid, hydrogen peroxide and bacteriocins, regulate vaginal pH levels and inhibit pathogenic colonization (2).
Bacterial vaginosis (BV) is caused by dysbiosis, or an imbalance in the vaginal microbiota. This condition is characterized by a decline in Lactobacillus abundance and an overgrowth of pathogens, such as Gardnerella vaginalis, Atopobium vaginae, Megasphaera spp., Prevotella spp., and Sneathia spp. (3,4). These microbial imbalances underscore the importance of maintaining a healthy vaginal microbiota to prevent gynecologic complications and reduce the transmission of sexually transmitted infections.
Of note, two other prevalent vaginal illnesses are candidiasis caused by Candida albicans and trichomoniasis, which is caused by Trichomonas vaginalis (5). Until recently, the endometrium was considered to be a sterile environment. A functional microbiome is present in the endometrium in physiological settings. Some research indicates that Lactobacillus is the major and representative genus of a healthy endometrium, while other research suggests different bacterial genera (6). After analyzing endometrial samples, Franasiak et al (7) found that the most prevalent genera were Lactobacillus and Flavobacterium. Lactobacillus (71.1%) was the most common bacteria found in endometrial fluid samples from fertile women, followed by Gardnerella, Bifidobacterium, Streptococcus and Prevotella (8).
One of the most common types of gynecological cancer is endometrial cancer (EC), which is highly related to the endocrine system (9). Numerous variables, including environmental circumstances, genetic susceptibility, hormonal imbalances (particularly involving estrogen and progesterone), heavy periods, being overweight, or being at an advanced age, may contribute to the development of EC (10). For instance, previous studies have demonstrated that the vaginal microbiota may play a role in the development of EC by directly interacting with endometrial tissue that is susceptible to the disease or by generating metabolites and inflammatory molecules that affect the course of cancer (11,12). Atopobium vaginae and Porphyromonas, among other bacteria that raise the vaginal pH levels, are more prevalent in the vaginal flora of women with endometrial hyperplasia or EC (13). This is considered to promote chronic endometrial inflammation that stimulates the carcinogenesis process (14). Therefore, the aim of the present study was to perform a comparison of the semi-quantitative and qualitative dynamics between the vaginal microbiota in women with vaginitis and EC in Sulaymaniyah, Iraq using next-generation sequencing (NGS). The present study focused on EC, excluding other cancer types.
Patients and methods
Sample collection
A total of 100 high vaginal swab samples were collected from Al-Sulaymaniyah Governorate, Iraq at Sulaimani Teaching Hospital, Sulaimaniyah, Iraq and Uper Arbat Health Center, Sulaimaniyah, Iraq, between September, 2022 and July, 2023. A total of 48 various types of gynecological cancer samples were collected and diagnosed through clinical findings and a histological examination; 16/58 samples were excluded due to negative pathohistological results. The ages of the women with gynecological cancers ranged from 18 to 83 years; there was a total number of 42 cancer cases. Of these, 16 cases were EC. Simultaneously, 42 other samples were collected from females with vaginitis, aged 21 to 66 years.
The College of Science Ethics Committee, College of Science, University of Baghdad, approved the research proposal in the present study. The College of Science Ethics committee expects to be informed about the study's progress, any serious adverse events occurring during the study, any revision in the protocol, and patient information/written informed consent and ask to be provided with a copy of the final report.
Molecular detection of vaginal swabs: Genomic DNA extraction
Genomic DNA was extracted from 6 vaginal specimens, including 2 cases of EC (D79-S27 and D80-S28) and 4 specimens from patients with vaginitis (D11-S18, D13-S19, D14-S20 and D65-S25) using the BIORON GmbH kit [BIORON Genomic DNA Mini kit (for Cells and Tissues) cat. no. 112005]. The selected samples depended on the similarity in some parameters (data are not published now) to reduce the difference as much as possible among the tested samples. These DNA samples were used to detect the diversity of the vaginal microbiome from patients with EC and vaginitis by sequencing the 16S rRNA gene using the iSeq method.
Measuring the DNA concentration and purity
DNA concentrations of the extracted samples were measured using the QuantiFluor dsDNA System kit from (cat. no. E2670, Promega Corporation) and analyzed using a Quantus fluorometer device (cat. no. E6150, Promega Corporation). The Quantus fluorometer yielded the following concentrations for the 4 infected subjects: (90, 103, 100 and 124 ng/µl). The concentrations for the 2 samples from patients with EC were 125 and 123 ng/µl.
Primer preparation
The present study used a primer set to identify the V3-V4 region of 16SrRNA (forwardD, 5'-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3' and reverse, 5'-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3'). Primers were synthesized and prepared by Macrogen, Inc., delivered in lyophilized form and reconstituted to a working concentration with nuclease-free water (Table I).
Library purification
The 16S ribosomal RNA gene amplicons for the Illumina MiSeq System kit were used to PCR amplify the 16S libraries using primers targeting the 16S rRNA gene hypervariable V3 and V4 regions with overhang adapter sequences (15). The thermal amplification profile included an initial denaturation at 95˚C for 3 min, followed by 8 cycles of 95˚C for 30 sec, 55˚C for 30 sec and 72˚C for 30 sec, with a final extension at 72˚C for 5 min and a hold at 4˚C. Following PCR amplification preparation, agarose gel electrophoresis was used for PCR amplicon purification, sample preparation, tiny fragment removal and dual-size DNA fragment selection.
NGS
In NGS, and according to the manufacturer's recommendations (Illumina, Inc.), the amplicons were pooled in equimolar concentrations of 100 pM for iSeq sequencing. Pooled libraries of dsDNA were diluted with 10 mM Tris (pH 8.5) solution to a concentration of 1 nM. Subsequently, 15 µl pooled dsDNA libraries were diluted with 85 µl (resuspension buffer) RSB volume to yield a 150 pM loading concentration. A total of 20 µl of the library containing 14 pooled indexed samples was loaded into the iSeq 100 system. Sequencing was performed using iSeq 100 i1 Reagent v2 (300-cycle) manufactured by Illumina, Inc. The datasets used and analyzed during the current study are available from the corresponding author at NCBI and the accession no. PRJNA1222970 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1222970).
Statistical analysis
All were analyzed using IBM SPSS Statistics for Windows, version 26.0. (IBM Corp.). The results were analyzed using the Chi-squared test and Fisher's exact test to compare the means of the parameters where necessary. A P-value <0.05 was considered to indicate a statistically significant difference.
Results and Discussion
Types and prevalence of gynecological cancer among the patients
Of the 58 patients with gynecological cancers histologically examined, 16 suspected cases were excluded following a histological examination; thus, 42 different gynecological cancer cases remained to be included in the present study, along with an additional 42 cases of vaginitis. The results of the histological diagnosis of the 42 gynecological cancer specimens revealed that only 16 (38.09%) patients with an age range between (30-67 years) and a mean age of 49.68 years had EC, 14 (33.33%) cases had ovarian cancer and 8 (19.05%) cases had polyps; in addition, there were 1 cases each of cervical cancer, fallopian tube cancer, endometrial-ovarian cancer and endometrial-ovarian-fallopian tube cancer.
The findings of the present study are in contrast to those of a recent study by Priyadarshini et al (16), which found that uterine cancer was the third most common type of cancer. As a result, various poorly known, diverse causes may likely play a role in these events (17).
Within the same framework, the study by Knudsen et al (18) from 2014 elucidated that the elderly had a potentially reduced survival rate and an age-dependent mortality rate for gynecological cancers. Additionally, Piechocki et al (19) noted an increase in the incidence of breast cancer, ovarian and uterine cancer, and a decrease in the incidence of cervical and vaginal cancer in Poland. The study by Somasegar et al (20) conducted in the United States analyzed trends in the mortality rates of patients with uterine cancer over a period of 50 years, with an emphasis on age and race and ethnicity. They suggested that the incidence of uterine cancer was 6-fold higher in women aged ≥70 years than in those aged 50 to 59 years. According to Duska et al (21), elderly individuals typically experience poorer outcomes and are diagnosed at a later stage. Thus, one of the primary risk factors for cancer is an older age.
Molecular identification of the vaginal microbiome: Detection of 16S rRNA gene in the vaginal microbiome using a PCR template
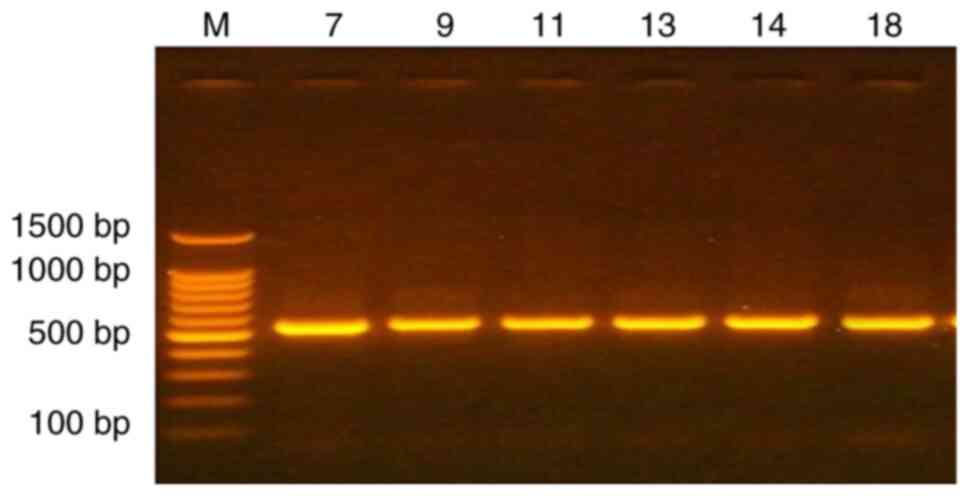
A total of 6 vaginal specimens were selected to amplify the conserved region of the thermo-stable nuclease that encodes for the 16S rRNA gene using universal primers. To detect the genotype of bacterial communities in vaginal swabs, the good quality and concentration DNA samples that were extracted from the vaginal specimens of patients with gynecological cancers, including EC and vaginitis, were used to confirm the type of bacteria using the PCR technique. The primer set (Table I), was used to cover the V3-V4 region in the 16S rRNA gene. This region is known to have higher resolution for low-rank taxa (bacteria and archaea) (22). The results presented in Fig. 1 revealed specific 550 bp bands, indicating the correct amplicon.
The human microbiome is anticipated to function as a novel and beneficial tool for classifying human epithelial materials, as Yao et al (23) found in 2021. Vaginal secretions are the most common biological specimen in several sexual assault cases; the characterization of these fluids is crucial to accurately determining the nature of the case. Knowledge of vaginal microbiota across different regions, physiological conditions and ethnic populations is constantly being improved. The present study performed high-throughput sequencing of the V3-V4 hypervariable regions of the 16S rRNA gene of vaginal samples collected from patients in the Al-Sulaymaniyah region, Iraq, who had EC and vaginitis to obtain comparable information about the vaginal microflora and its association with cancer sequelae in women.
Illumina iSeq sequencing for the identification of bacteria in the vaginal microbiome
A total of 6 DNA samples were extracted from the patients with gynecological cancers, including 2 cases of EC (NCBI Accession nos. SRX27671438 and SRX27671439) another 4 cases of vaginitis (NCBI Accession nos. SRX27671443, SRX27671442, SRX27671444 and SRX27671448) used as an indicator for vaginal microbiome, these data have been deposited in (NCBI). The DNA samples were sent to Macrogen Inc. to identify the bacterial community among these groups by NGS (iSeq sequencing). All DNA samples used in this experiment were passed through the quantification analysis.
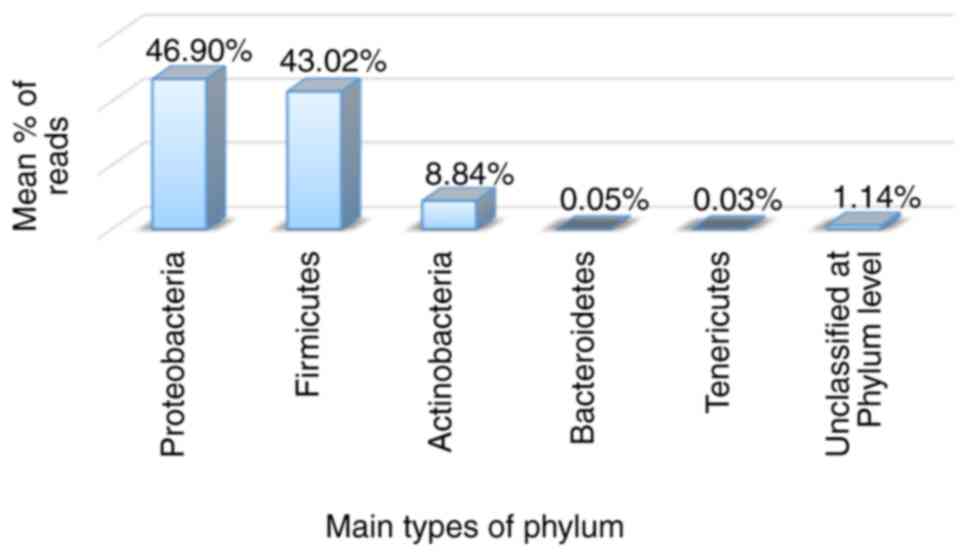
One of the primary techniques in studying the taxonomy of the microbiome community is the (16S rRNA). It plays a crucial role in the survival and functions of the cell, and it is considered a golden standard in microbial genotyping. The 16S rRNA gene is comprise of two hypervariable regions bordered by more conserved sequences. Microbial diversification studies typically employ the dual-indexed custom primer 16S rRNA gene sequencing technology for the V4 hypervariable region. This region was used by the Human Microbiome Project to provide adequate information for the taxonomic classification of microbial communities obtained from specimens related to human microbiome research. However, the approach outlined could be extended to any location (24,25), yet it was designed for iSeq sequencing. Griffin et al (26) demonstrated the utility of NGS for resolving polyploid complexes. The advantage of this technique is discovering the low-abundant microbe. It allows covering large numbers of reads with high depth in a single experiment. The mean percentages of bacterial phyla for the 4 patients in the present study suffering from different types of vaginosis are presented in Fig. 2.
A total of iSeq 51,459 reads were created in the present study and were used for downstream analyses (Fig. 2). The reads were classified into 32 operational taxonomic units (OTUs) representing the individual bacterial species. The most abundant phyla detected in the present study were Proteobacteria (46.90%), Firmicutes (43.02%), Actinobacteria (8.84%), Bacteroidetes (0.05%), Tenericutes (0.03%) and unclassified at the phylum level (1.14%). The values in parentheses represent the average percentage of bacteria at a specific taxonomic level in the vaginitis cases. As demonstrated in Fig. 2, the most dominant phyla are Proteobacteria and Firmicutes; these findings are in accordance with those in the study by Sroka-Oleksiak et al (27) who also revealed 4 phyla: Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes. It can be suggested that the differences between the percentages of the two studies may be related to the disruptions among vaginal microbiomes as a result of infections, as previously mentioned by Srinivasan et al (28).
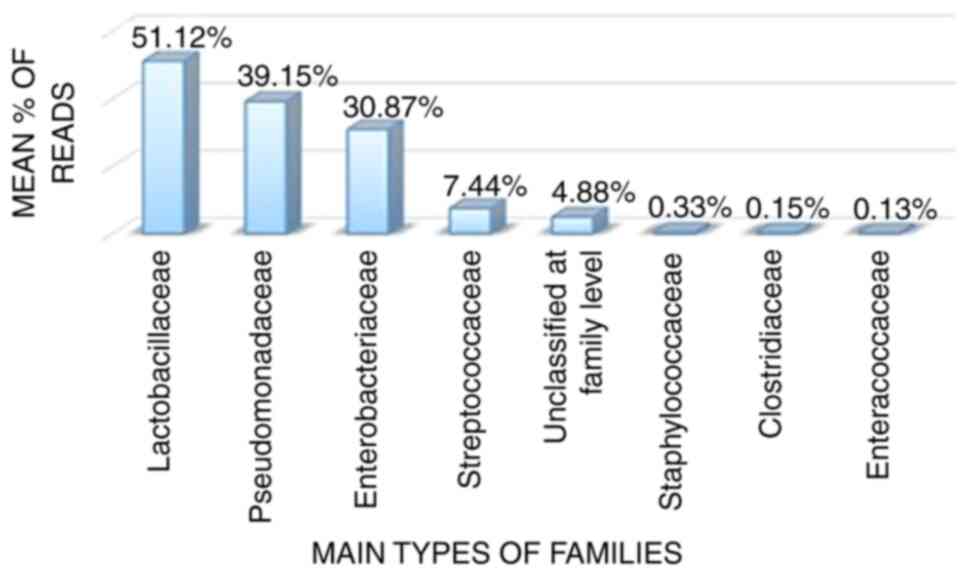
The results presented in Fig. 3 revealed that the most predominant bacterial families, in descending order, were as follows: Lactobacillaceae (51.12%), Pseudomonas (39.15%), Enterobacteriaceae (30.87%), Streptococcaceae (7.44%), Staphylococcaceae (0.33%), Clostridiaceae (0.15%), Enterococcaceae (0.13%) and others under unclassified at family level (4.88%).
The results of the present study were completely incompatible with those documented in the study by Srinivasan et al (29), which revealed that ~20% of pregnant women may suffer from BV, the most prevalent vaginal condition in patients. In BV, the overgrowth of characteristically non-Lactobacillus anaerobic bacteria, such as Mobiluncus spp., Gardnerella vaginalis, and Atopobium vaginae, leads to a disruption of the ecosystem of vaginal balance and the modification of the vaginal milieu, which may result in developing clinical signs, such as vaginal discharge and itchiness. Nevertheless, almost 50% of the patients who have BV are asymptomatic or have fewer clinical signs. However, Yao et al (23) demonstrated that there was minimal variation in the makeup of the main bacteria in the vagina, which were primarily Lactobacillus and Gardnerella species.
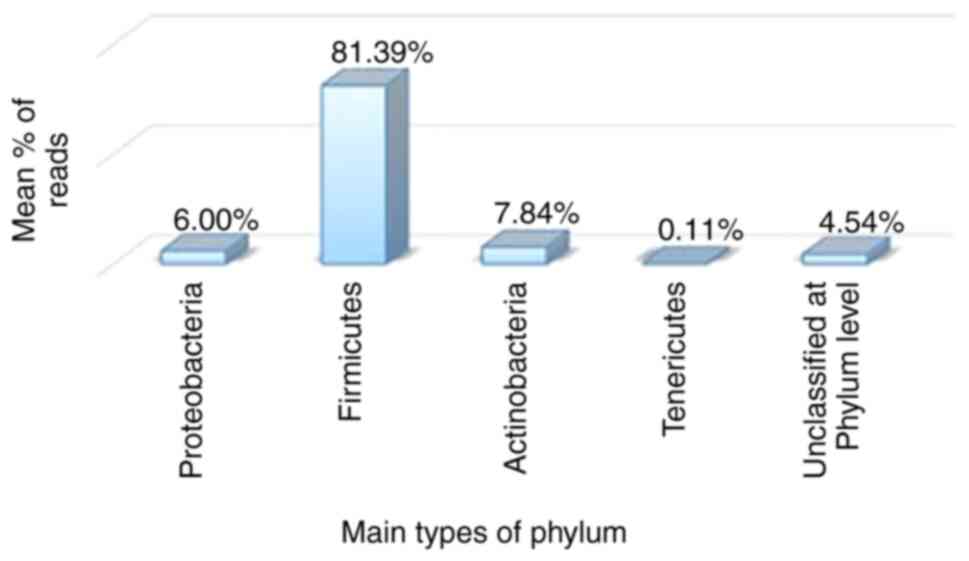
In the same context, Fig. 4 illustrates the mean percentages of bacterial phyla for 2 patients with EC. A total of iSeq 23,817 reads were created in the present study and were used for downstream analyses (Fig. 4). The reads were classified into 16 OTUs representing the individual bacterial species. The most abundant phyla detected in the present study were Firmicutes (81.39%), Actinobacteria (7.84%), Proteobacteria (6.00%), unclassified at the phylum level (4.54%) and Tenericutes (0.11%); these percentages represent the average percentage of bacteria at a specific taxonomic level in the endometrial cancer cases. From these phyla, as illustrated in Fig. 4, the phylum, Firmicutes, was predominant in EC, as also demonstrated by Lee et al (30), who documented that vaginal microbiota are related to the health of the reproductive system of women. When Lactobacillus levels are reduced, the majority of the vaginal microbiota tend to become imbalanced, which can lead to the development of bacterial vaginosis (31). Srinivasan et al (28) reported that there are several clinical signs related to vaginal infection, such as a vaginal pH >4.5, the presence of clue cells, amine odor under the whiff test, and an increased thin homogenous vaginal discharge linked with specific microorganisms such as Atopobium vaginae, Gardnerella vaginalis, and Leptotrichia amnionii. Tamrakar et al (32) indicated that the Lactobacillus iners is considered a marker of vaginal microbiome disorder as its prevalence has been linked with that of other bacterial vaginosis-related bacteria, such as Leptotrichia, Eggerthella, Megasphaera and bacterial vaginosis.
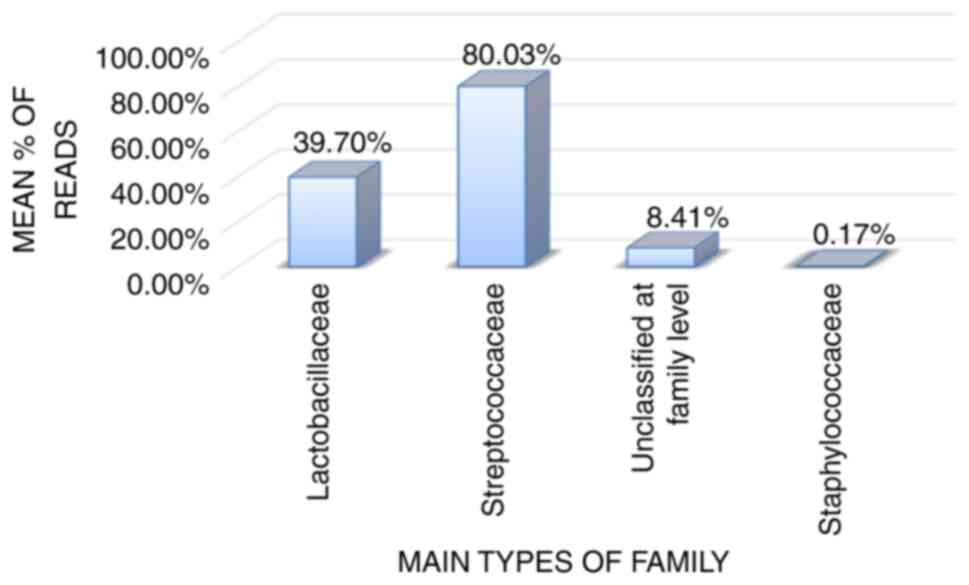
The results shown in Figs. 2 and 4 yielded significantly differences P<0.05). Based on the phylum classification distributions, the results are statistically significant, indicating an association between endometrial bacterial sample disorders and vaginitis. The observed differences show substantial variations in the distribution of phyla between the two cases (Table II). The observed comparable statistical analysis revealed that a P-value of 0.001, indicating a highly significant difference in the family classification distribution between the two conditions [vaginitis (Fig. 3) and EC (Fig. 5); Table III]. Based on the data presented in Fig. 5, the most common bacterial families were Streptococcaceae (80.03%), Lactobacillaceae (39.70%) and Staphylococcaceae (0.17%). The remaining taxa (8.41%) are not categorized at the family level. A P-value of 0.001 suggests that the family categorization distributions for EC (Fig. 5) and vaginitis (Fig. 3) cases differ significantly.
Table IIChi-squared test analysis for the distribution of bacterial phyla between the vaginitis and endometrial cancer samples. |
Table IIIFisher's exact test analysis for the distribution of bacterial Families between the vaginitis and endometrial cancer samples. |
Moreno and Simon (33) demonstrated that the microbiome related to the reproductive tract comprised 9% of the human microbiota, which includes the flora in the vagina and endometrial follicular fluid in women. The composition of the vaginal microbiota depends on hormonal fluctuations, sexual behaviors, age and menstruation, as well as on the use of drugs such as antibiotics and probiotics that may lead to an imbalance. The results presented in Fig. 5 reveal the decrease in the Lactobacillaceae reading frequency compared to Streptococcaceae; these variations may be explained by Wan et al (34), who reported the association of hormonal factors and the development of EC. It is certain however, that excess levels of or unopposed estrogen are a major risk factor. Vajpeyee et al (35) documented that endometrial thickness had a significant impact on the levels of luteinizing hormone, estrogen and progesterone. In addition, microbial abundance alteration depends on the reproductive system states, regardless of the degree of pathogenicity that can affect fertilization, implantation and subsequent embryonic development. After comparing pregnant and non-pregnant patients, pregnant patients had a higher abundance of Firmicutes and Proteobacteria, and a lower abundance of Actinobacteria, Fusobacterium and Bacteroidetes at the phylum level (36). Therefore, it can be suggested that different sexual hormonal levels may directly or indirectly affect the vaginal microbiome (37).
In short, these samples were classified into 11 and 9 phyla, 12 and 9 classes, 18 and 11 orders, 20 and 14 families, and finally, 25 and 15 genera for the vaginitis and EC specimens, respectively. The most abundant bacterial families in the present study for the vaginitis samples were Lactobacillaceae, Staphylococcaceae, Streptococcaceae, Enterococcaceae and Clostridiaceae (Firmicutes phylum); Paenibacillaceae (Bacillota phylum); Pseudomonadaceae, Anaplasmataceae, Acetobacteraceae, Enterobacteriaceae, Desulfovibrionaceae, Vibrionaceae, Ferrimonadaceae and Shewanellaceae (Proteobacteria phylum); Bifidobacteriaceae, Mycobacteriaceae, Glycomycetaceae, Intrasporangiaceae (Actinobacteria phylum); Prevotellaceae (Bacteroidetes phylum); and others are unclassified phylum and family levels. In addition to Streptococcaceae, Staphylococcaceae, Planococcaceae, and Leuconostocaceae (Firmicutes phylum); Paenibacillaceae (Bacillota phylum); Brucellaceae, Bartonellaceae, (Proteobacteria phylum); Corynebacteriaceae, Intrasporangiaceae and Bifidobacteriaceae (Actinobacteria phylum); Desulfovibrionaceae (Thermodesulfobacteriota phylum); Phyllobacteriaceae (Pseudomonadota phylum); and others are unclassified at the phylum and family levels, which represent the most abundant families in endometrial samples.
Thus, nine bacterial phyla and eight families were identified as being shared between vaginitis and endometrial cancer. The shared phyla included Proteobacteria, Firmicutes, Actinobacteria, Tenericutes, Bacteroidetes, Verrucomicrobia, Fusobacteria and Spirochaetes, along with other unclassified taxa at the phylum level. Similarly, at the family level, the shared bacterial families comprised Lactobacillaceae, Streptococcaceae, Staphylococcaceae, Bifidobacteriaceae, Intrasporangiaceae, Desulfovibrionaceae and Paenibacillaceae, in addition to other unclassified families.
The present study identified nine shared phyla, classes, orders, and families between the two groups. Additionally, the presence of Lactobacillus does not necessarily suppress another vaginal microbiota. In addition, in each case, the vaginal microbiomes produce a microenvironment suitable for particular microbial growth that may affect directly or indirectly the ecosystem of the female reproductive system. Thus, it may highlight the need for further research to determine the real causes using larger sample sizes for potential therapeutic applications.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be found in the NCBI database under accession number PRJNA1222970 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1222970).
Authors' contributions
All authors (DFA, MAA and TAH) contributed significantly to the acquisition, analysis and interpretation of the data. MAA suggested the subject of research, and both DF and TAH collected samples from the hospital for analysis and presented accurate research results. All authors have reviewed and have read and approved the final version of the manuscript. DFA and MAA confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The College of Science, University of Baghdad Ethics Committee approved the research proposal in the present study. The College of Science Ethics committee expects to be informed about the study's progress, any serious adverse events occurring during the study, any revision in the protocol, and patient information/written informed consent and ask to be provided with a copy of the final report. None of the investigators and co-investigators participating in this study took part in the decision-making and voting procedure for this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Rosner J, Samardzic T and Sarao MS: Physiology, Female Reproduction. In: StatPearls. StatPearls Publishing, Treasure Island, FL, 2025. | |
|
Wu S, Hugerth LW, Schuppe-Koistinen I and Du J: The right bug in the right place: Opportunities for bacterial vaginosis treatment. NPJ Biofilms Microbiomes. 8(34)2022.PubMed/NCBI View Article : Google Scholar | |
|
Abbe C and Mitchell CM: Bacterial vaginosis: A review of approaches to treatment and prevention. Front Reprod Health. 5(1100029)2023.PubMed/NCBI View Article : Google Scholar | |
|
Moreno I and Franasiak JM: Endometrial microbiota-new player in town. Fertil Steril. 108:32–39. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Alwaily ER, Flaih MH, Abood MS and Hussein KR: Determination of fungal and parasitic infections caused vaginitis: Molecular identification of Candida parapsilosis in Al-Nasiriyah city, Iraq. Baghdad Sci J. (20)2023. | |
|
Punzón-Jiménez P and Labarta E: The impact of the female genital tract microbiome in women health and reproduction: A review. J Assist Reprod Genet. 38:2519–2541. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Franasiak JM, Werner MD, Juneau CR, Tao X, Landis J, Zhan Y, Treff NR and Scott RT: Endometrial microbiome at the time of embryo transfer: Next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet. 33:129–136. 2016.PubMed/NCBI View Article : Google Scholar | |
|
García-Velasco JA, Budding D, Campe H, Malfertheiner SF, Hamamah S, Santjohanser C, Schuppe-Koistinen I, Nielsen HS, Vieira-Silva S and Laven J: The reproductive microbiome-clinical practice recommendations for fertility specialists. Reprod Biomed Online. 41:443–453. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Mahdy H, Vadakekut ES and Crotzer D: Endometrial Cancer. In: StatPearls. StatPearls Publishing, Treasure Island, FL, 2025. Available from: https://www.ncbi.nlm.nih.gov/books/NBK525981. | |
|
Bassette E and Ducie JA: Endometrial cancer in reproductive-aged females: Etiology and pathogenesis. Biomedicines. 12(886)2024.PubMed/NCBI View Article : Google Scholar | |
|
Walsh DM, Hokenstad AN, Chen J, Sung J, Jenkins GD, Chia N, Nelson H, Mariani A and Walther-Antonio MRS: Postmenopause as a key factor in the composition of the Endometrial Cancer Microbiome (ECbiome). Sci Rep. 9(19213)2019.PubMed/NCBI View Article : Google Scholar | |
|
Al-Ruba'ei SHN, Jasim RK and Al-Rawaq KJS: Serum antioxidant status in Iraqi women with endometrial cancer. J Fac Med Baghdad. 58:378–383. 2016. | |
|
Lu W, He F, Lin Z, Liu S, Tang L, Huang Y and Hu Z: Dysbiosis of the endometrial microbiota and its association with inflammatory cytokines in endometrial cancer. Int J Cancer. 148:1708–1716. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Aniewski P, Ilhan ZE and Herbst-Kralovetz MM: The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. 17:232–250. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M and Glöckner FO: Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41(e1)2013.PubMed/NCBI View Article : Google Scholar | |
|
Priyadarshini S, Swain PK, Agarwal K, Jena D and Padhee S: Trends in gynecological cancer incidence, mortality, and survival among elderly women: A SEER study. Aging Med (Milton). 7:179–188. 2024.PubMed/NCBI View Article : Google Scholar | |
|
Siegel RL, Miller KD, Wagle NS and Jemal A: Cancer statistics. CA Cancer J Clin. 73:17–48. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Knudsen Ør A, Schledermann D, Nyvang GB, Mogensen O and Herrstedt J: On behalf of the Academy of Geriatric Cancer Research (Age Care). Trends in gynecologic cancer among elderly women in Denmark, 1980-2012. Acta Oncol. 55 (Suppl 1):S65–S73. 2016. | |
|
Piechocki M, Koziołek W, Sroka D, Matrejek A, Miziołek P, Saiuk N, Sledzik M, Jaworska A, Bereza K, Pluta E and Banas T: Trends in incidence and mortality of gynecological and breast cancers in Poland (1980-2018). Clin Epidemiol. 14:95–114. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Somasegar S, Bashi A, Lang SM, Liao CI, Johnson C, Darcy KM, Tian C, Kapp DS and Chan JK: Trends in uterine cancer mortality in the United States: A 50-year population-based analysis. Obstet Gynecol. 142:978–986. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Duska L, Shahrokni A and Powell M: Treatment of older women with endometrial cancer: Improving outcomes with personalized care. Am Soc Clin Oncol Educ Book. 36:164–174. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Hrovat K, Dutilh BE, Medema MH and Melkonian C: Taxonomic resolution of different 16S rRNA variable regions varies strongly across plant-associated bacteria. ISME Commun. 4(ycae034)2024.PubMed/NCBI View Article : Google Scholar | |
|
Yao T, Wang Z, Liang X, Liu C, Yu Z, Han X, Liu R, Liu Y, Liu C and Chen L: Signatures of vaginal microbiota by 16S rRNA gene: potential bio-geographical application in Chinese Han from three regions of China. Int J Legal Med. 135:1213–1224. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Kozich JJ, Westcott SL, Baxter NT, Highlander SK and Schloss PD: Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 79:5112–5120. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Voelkerding KV, Dames SA and Durtschi JD: Next-generation sequencing: From basic research to diagnostics. Clin Chem. 55:641–658. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Griffin PC, Robin C and Hoffmann AA: A next-generation sequencing method for overcoming the multiple gene copy problem in polyploid phylogenetics, applied to Poa grasses. BMC Biol. 9(19)2011.PubMed/NCBI View Article : Google Scholar | |
|
Sroka-Oleksiak A, Gosiewski T, Pabian W, Gurgul A, Kapusta P, Ludwig-Słomczyńska AH, Wołkow PP and Brzychczy-Włoch M: Next-generation sequencing as a tool to detect vaginal microbiota disturbances during pregnancy. Microorganisms. 8(1813)2020.PubMed/NCBI View Article : Google Scholar | |
|
Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL and Hall RW: Bacterial communities in women with bacterial vaginosis: High resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One. 7(e37818)2012.PubMed/NCBI View Article : Google Scholar | |
|
Srinivasan S, Morgan MT, Fiedler TL, Djukovic D, Hoffman NG, Raftery D, Marrazzo JM and Fredricks DN: Metabolic signatures of bacterial vaginosis. mBio. 6:e00204–15. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Lee S, Oh KY, Hong H, Jin CH, Shim E, Kim SH and Kim BY: Community state types of vaginal microbiota and four types of abnormal vaginal Microbiota in Pregnant Korean Women. Front Public Health. 8(507024)2020.PubMed/NCBI View Article : Google Scholar | |
|
Alshammery RM and Alaubydi MA: Extraction and partial purification of Exo-polysaccharide from Lactobacillus rhamnosus isolated from vaginal specimens. Iraqi J Sci. 61:754–764. 2020. | |
|
Tamrakar R, Yamada T, Furuta I, Cho K, Morikawa M, Yamada H, Sakuragi N and Minakami H: Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women. BMC Infect Dis. 7(128)2007.PubMed/NCBI View Article : Google Scholar | |
|
Moreno I and Simon C: Deciphering the effect of reproductive tract microbiota on human reproduction. Reprod Med Biol. 18:40–50. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Wan J, Gao Y, Zeng K, Yin Y, Zhao M, Wei J and Chen Q: The levels of the sex hormones are not different between type 1 and type 2 endometrial cancer. Sci Rep. 6(39744)2016.PubMed/NCBI View Article : Google Scholar | |
|
Vajpeyee M, Tiwari S, Yadav LB and Tank P: Assessment of bacterial diversity associated with assisted reproductive technologies through next-generation sequencing. Middle East Fertility Society J: Sep 5, 2022. doi: 10.1186/s43043-022-00117-3. | |
|
Smid MC, Ricks NM, Panzer A, Mccoy AN, Azcarate-Peril MA, Keku TO and Boggess KA: Maternal gut microbiome biodiversity in pregnancy. Am J Perinatol. 35:24–30. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Al-Sarray SA, Al Aubydi MA and Muhsin SM: Serum caspase 3 and lysozyme level in abortive Iraqi Women Infected with Toxoplasma gondii and urinary tract infections. Indian J Public Health Res Dev: 10, 2019. |