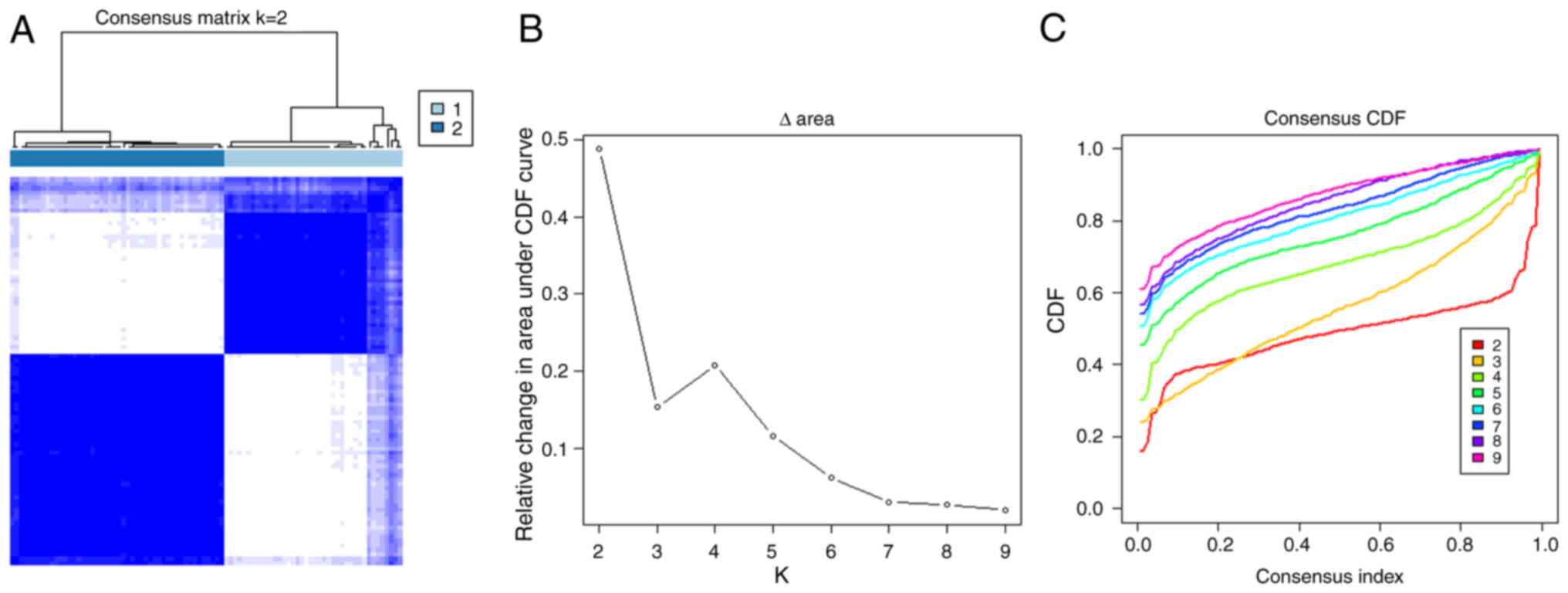
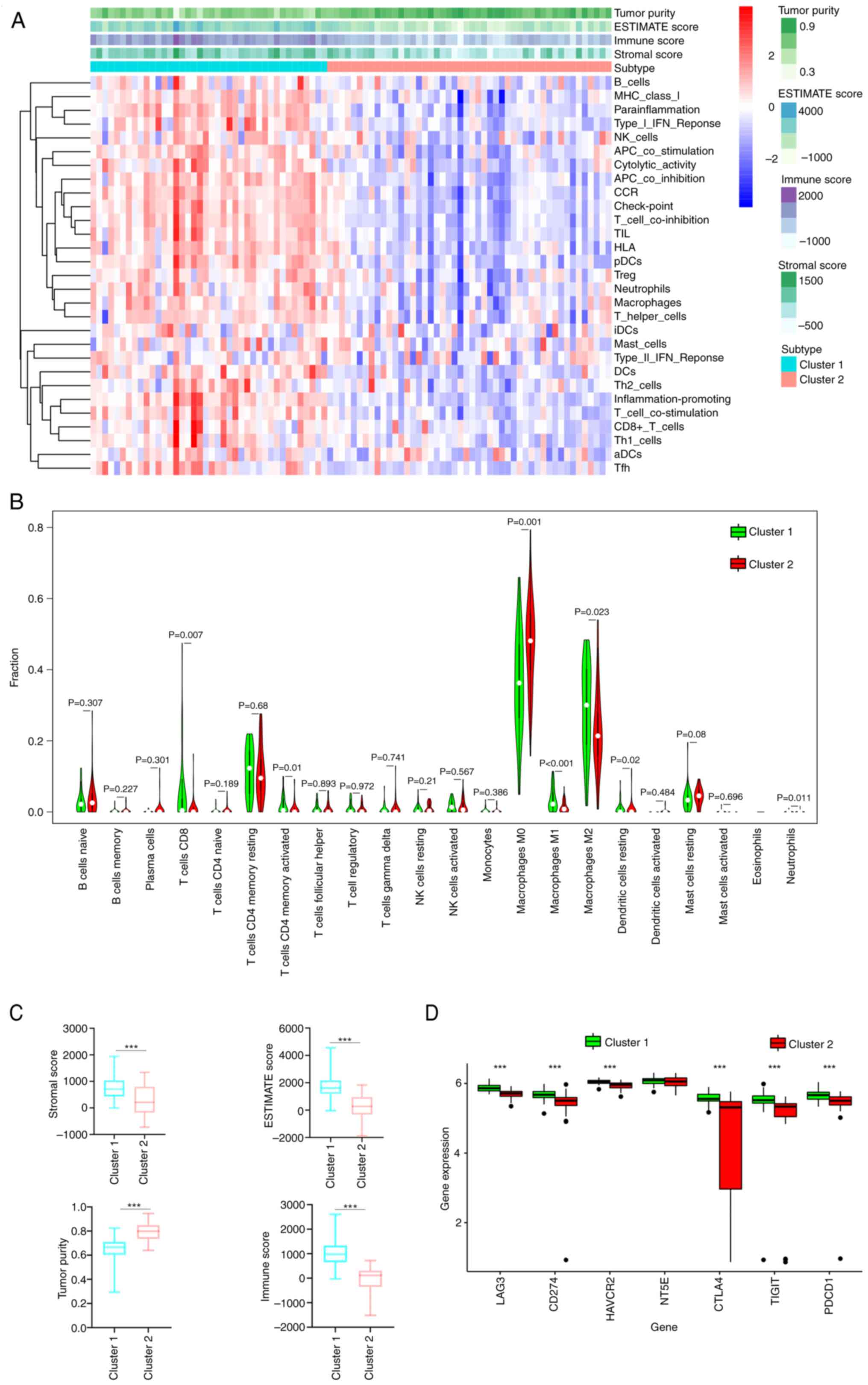
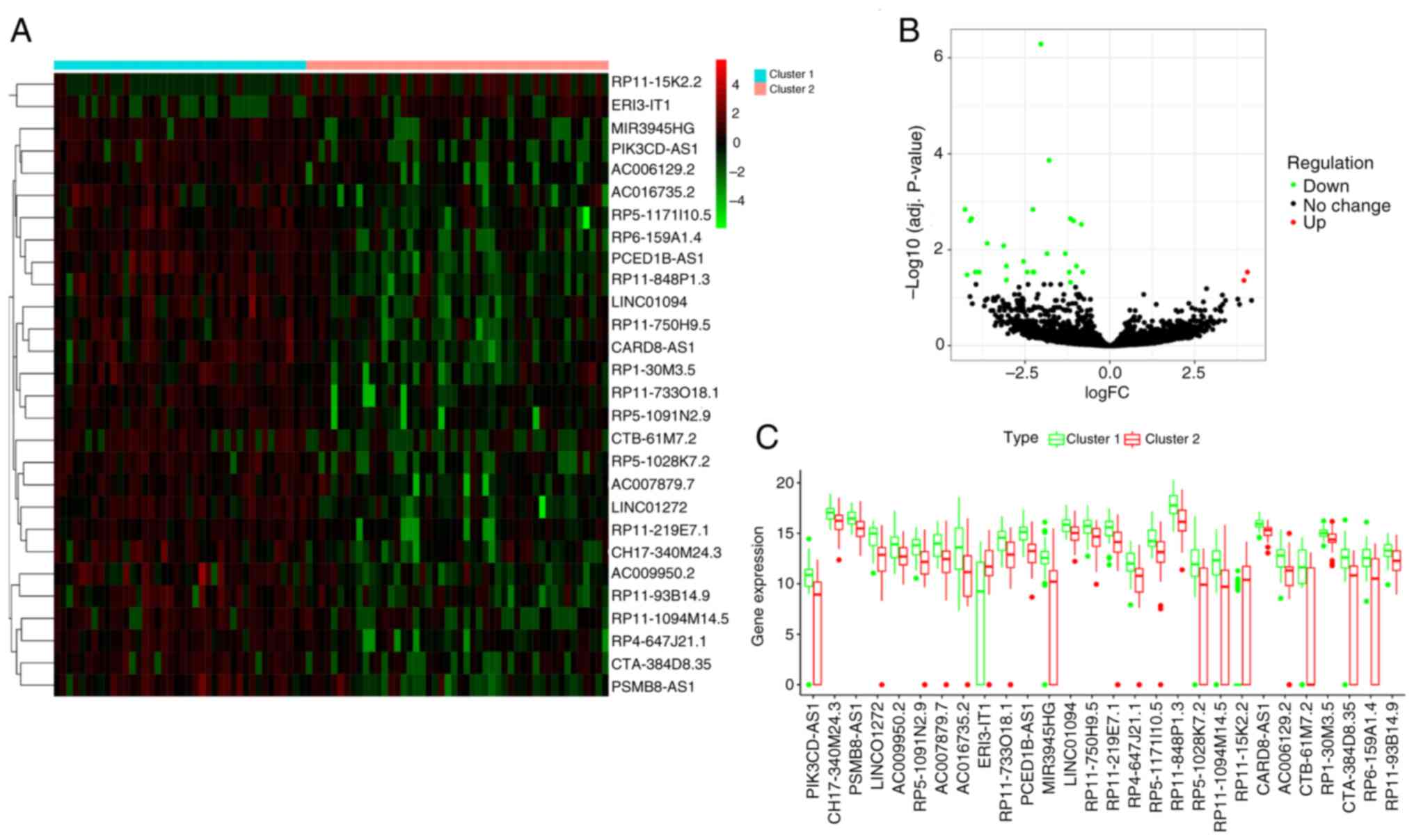
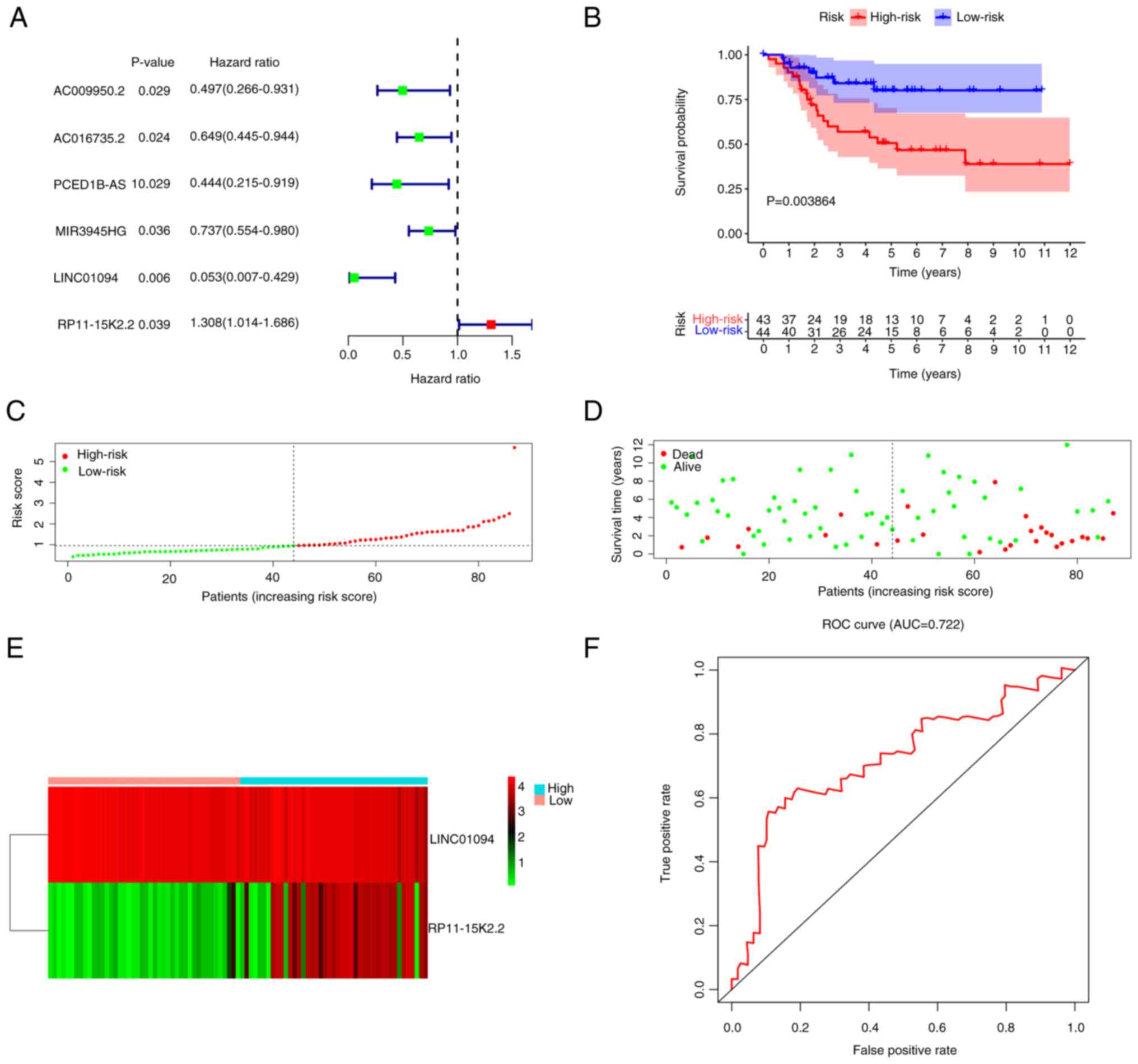
|
1
|
Rothzerg E, Xu J and Wood D: Different
subtypes of Osteosarcoma: histopathological patterns and clinical
behaviour. J Mol Pathol. 4:99–108. 2023. View Article : Google Scholar
|
|
2
|
Moukengue B, Lallier M, Marchandet L,
Baud'huin M, Verrecchia F, Ory B and Lamoureux F: Origin and
therapies of osteosarcoma. Cancers (Basel). 14:35032022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gaspar N, Hung GY, Strauss SJ,
Campbell-Hewson Q, Dela Cruz FS, Glade Bender JL, Koh KN, Whittle
SB, Chan GC, Gerber NU, et al: Lenvatinib plus ifosfamide and
etoposide in children and young adults with relapsed osteosarcoma:
A phase 2 randomized clinical trial. JAMA Oncol. 10:1645–1653.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
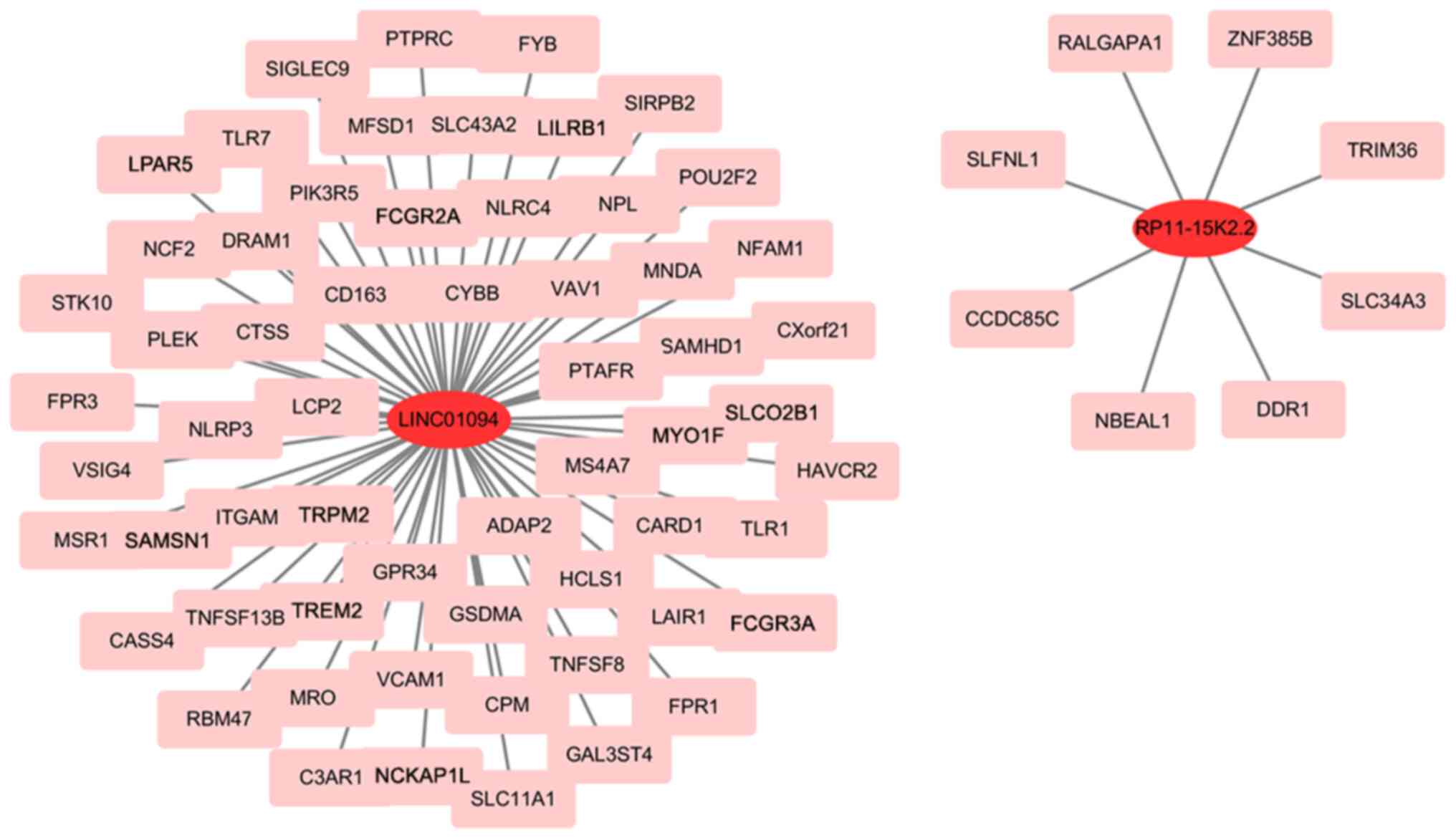
Pei Y, Yao Q, Li Y, Zhang X and Xie B:
microRNA-211 regulates cell proliferation, apoptosis and
migration/invasion in human osteosarcoma via targeting EZRIN. Cell
Mol Biol Lett. 24:482019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen JK, Cote GM, Choy E, Yang P, Harmon
D, Schwab J, Nielsen GP, Chebib I, Ferrone S, Wang X, et al:
Programmed cell death ligand 1 expression in osteosarcoma. Cancer
Immunol Res. 2:690–698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
D'Angelo SP, Mahoney MR, Van Tine BA,
Atkins J, Milhem MM, Jahagirdar BN, Antonescu CR, Horvath E, Tap
WD, Schwartz GK and Streicher H: Nivolumab with or without
ipilimumab treatment for metastatic sarcoma (Alliance A091401): Two
open-label, non-comparative, randomised, phase 2 trials. Lancet
Oncol. 19:416–426. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie L, Liang X, Xu J, Sun X, Liu K, Sun K,
Li Y, Tang X, Li X, Zhan X, et al: Exploratory study of an
anti-PD-L1/TGF-β antibody, TQB2858, in patients with refractory or
recurrent osteosarcoma and alveolar soft part sarcoma: A report
from Chinese sarcoma study group (TQB2858-Ib-02). BMC Cancer.
23:8682023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tawbi HA, Burgess M, Bolejack V, Van Tine
B, Schuetze S, Hu J, D'Angelo S, Attia S, Riedel RF, Priebat DA, et
al: Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma
(SARC028): A multicentre, two-cohort, single-arm, open-label, phase
2 trial. Lancet Oncol. 18:1493–1501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pardoll DM: The blockade of immune
checkpoints in cancer immunotherapy. Nat Rev Cancer. 12:252–264.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Daud AI, Wolchok JD, Robert C, Hwu WJ,
Weber JS, Ribas A, Hodi FS, Joshua AM, Kefford R, Hersey P, et al:
Programmed death-ligand 1 expression and response to the
anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin
Oncol. 34:4102–4109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhai X, Zhao J, Wang Y, Wei X, Li G, Yang
Y, Chen Z, Bai Y, Wang Q, Chen X and Li M: Bibliometric analysis of
global scientific research on lncRNA: A swiftly expanding trend.
Biomed Res Int. 2018:76250782018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY,
Liu Q, Zhao G and Zhang ZZ: The lncRNA UCA1 promotes proliferation,
migration, immune escape and inhibits apoptosis in gastric cancer
by sponging anti-tumor miRNAs. Mol Cancer. 18:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li C, Hu J, Hu X, Zhao C, Mo M, Zu X and
Li Y: LncRNA SNHG9 is a prognostic biomarker and correlated with
immune infiltrates in prostate cancer. Transl Androl Urol.
10:215–226. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tu J, Wu F, Chen L, Zheng L, Yang Y, Ying
X, Song J, Chen C, Hu X, Zhao Z and Ji J: Long non-coding RNA PCAT6
induces M2 polarization of macrophages in cholangiocarcinoma
via modulating miR-326 and RhoA-ROCK signaling pathway.
Front Oncol. 10:6058772021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nguyen CB, Kumar S, Zucknick M, Kristensen
VN, Gjerstad J, Nilsen H and Wyller VB: Associations between
clinical symptoms, plasma norepinephrine and deregulated immune
gene networks in subgroups of adolescent with chronic fatigue
syndrome. Brain Behav Immun. 76:82–96. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Doncheva NT, Morris JH, Gorodkin J and
Jensen LJ: Cytoscape stringApp: Network analysis and visualization
of proteomics data. J Proteome Res. 18:623–632. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang Z, Jiang J, Zhao B, Yang H, Wang Y,
Guo S, Deng Y, Lu D, Ma T, Wang H and Wang J: CPNE1 silencing
inhibits the proliferation, invasion and migration of human
osteosarcoma cells. Oncol Rep. 39:643–650. 2018.PubMed/NCBI
|
|
22
|
Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang
S and Liu X: Long noncoding RNA MALAT1 as a potential therapeutic
target in osteosarcoma. J Orthop Res. 34:932–941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang SD, Li HY, Li BH, Xie T, Zhu T, Sun
LL, Ren HY and Ye ZM: The role of CTLA-4 and PD-1 in anti-tumor
immune response and their potential efficacy against osteosarcoma.
Int Immunopharmacol. 38:81–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsukahara T, Emori M, Murata K, Mizushima
E, Shibayama Y, Kubo T, Kanaseki T, Hirohashi Y, Yamashita T, Sato
N and Torigoe T: The future of immunotherapy for sarcoma. Expert
Opin Biol Ther. 16:1049–1057. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scott MC, Temiz NA, Sarver AE, Larue RS,
Rathe SK, Varshney J, Wolf NK, Moriarity BS, O'Brien TD, Spector
LG, et al: Comparative transcriptome analysis quantifies immune
cell transcript levels, metastatic progression and survival in
osteosarcoma. Cancer Res. 78:326–337. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Merchant MS, Melchionda F, Sinha M, Khanna
C, Helman L and Mackall CL: Immune reconstitution prevents
metastatic recurrence of murine osteosarcoma. Cancer Immunol
Immunother. 56:1037–1046. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu H, Wang X, Wu J, Ji H, Chen Z, Guo H
and Hou J: Long non-coding RNA LINC01094 promotes the development
of clear cell renal cell carcinoma by upregulating SLC2A3 via
microRNA-184. Front Genet. 11:5629672020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li XX and Yu Q: Linc01094 accelerates the
growth and metastatic-related traits of glioblastoma by sponging
miR-126-5p. Onco Targets Ther. 13:9917–9928. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu J, Zhang P, Sun H and Liu Y:
LINC01094/miR-577 axis regulates the progression of ovarian cancer.
J Ovarian Res. 13:1222020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kotiyal S and Bhattacharya S: Breast
cancer stem cells, EMT and therapeutic targets. Biochem Biophys Res
Commun. 453:112–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lou Y, Diao L, Cuentas ER, Denning WL,
Chen L, Fan YH, Byers LA, Wang J, Papadimitrakopoulou VA, Behrens
C, et al: Epithelial-mesenchymal transition is associated with a
distinct tumor microenvironment including elevation of inflammatory
signals and multiple immune checkpoints in lung adenocarcinoma.
Clin Cancer Res. 22:3630–3642. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yi Q, Zhu G, Zhu W, Wang J, Ouyang X, Yang
K, Fan Y and Zhong J: LINC01094: A key long non-coding RNA in the
regulation of cancer progression and therapeutic targets. Heliyon.
10:e375272024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang F, Wang M, Shi J and Xu G: IncRNA
MALAT1 regulates the proliferation, apoptosis, migration, and
invasion of osteosarcoma cells by targeting miR-873-5p/ROCK1. Crit
Rev Eukaryot Gene Expr. 33:67–79. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu M, Liu C, Li X and Li S: RP11-79H23.3
inhibits the proliferation and metastasis of non-small-cell lung
cancer through promoting miR-29c. Biochem Genet. 61:506–520. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Lv H, Liu X, Xu L, Li T, Zhou H,
Zhu H, Hao C, Lin C and Zhang Y: The RP11-417E7.1/THBS2 signaling
pathway promotes colorectal cancer metastasis by activating the
Wnt/β-catenin pathway and facilitating exosome-mediated M2
macrophage polarization. J Exp Clin Cancer Res. 43:1952024.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li L, Ouyang Y, Wang W, Hou D and Zhu Y:
The landscape and prognostic value of tumor-infiltrating immune
cells in gastric cancer. PeerJ. 7:e79932019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gu-Trantien C, Loi S, Garaud S, Equeter C,
Libin M, de Wind A, Ravoet M, Le Buanec H, Sibille C,
Manfouo-Foutsop G, et al: CD4+ follicular helper T cell
infiltration predicts breast cancer survival. J Clin Invest.
123:2873–2892. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gambardella V, Castillo J, Tarazona N,
Gimeno-Valiente F, Martínez-Ciarpaglini C, Cabeza-Segura M, Roselló
S, Roda D, Huerta M, Cervantes A and Fleitas T: The role of
tumor-associated macrophages in gastric cancer development and
their potential as a therapeutic target. Cancer Treat Rev.
86:1020152020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Harrington BK, Wheeler E, Hornbuckle K,
Shana'ah AY, Youssef Y, Smith L, Hassan Q II, Klamer B, Zhang X,
Long M, et al: Modulation of immune checkpoint molecule expression
in mantle cell lymphoma. Leuk Lymphoma. 60:2498–2507. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wolchok JD, Kluger H, Callahan MK, Postow
MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K,
et al: Nivolumab plus ipilimumab in advanced melanoma. N Engl J
Med. 369:122–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Motzer RJ, Rini BI, Mcdermott DF, Redman
BG, Kuzel TM, Harrison MR, Vaishampayan UN, Drabkin HA, George S,
Logan TF, et al: Nivolumab for metastatic renal cell carcinoma:
Results of a randomized phase II trial. J Clin Oncol. 33:1430–1437.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Goralski KB, Jackson AE, McKeown BT and
Sinal CJ: More than an adipokine: The complex roles of chemerin
signaling in cancer. Int J Mol Sci. 20:47782019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rodriguez GM, Bobbala D, Serrano D, Mayhue
M, Champagne A, Saucier C, Steimle V, Kufer TA, Menendez A,
Ramanathan S and Ilangumaran S: NLRC5 elicits antitumor immunity by
enhancing processing and presentation of tumor antigens to CD8(+) T
lymphocytes. Oncoimmunology. 5:e11515932016. View Article : Google Scholar : PubMed/NCBI
|