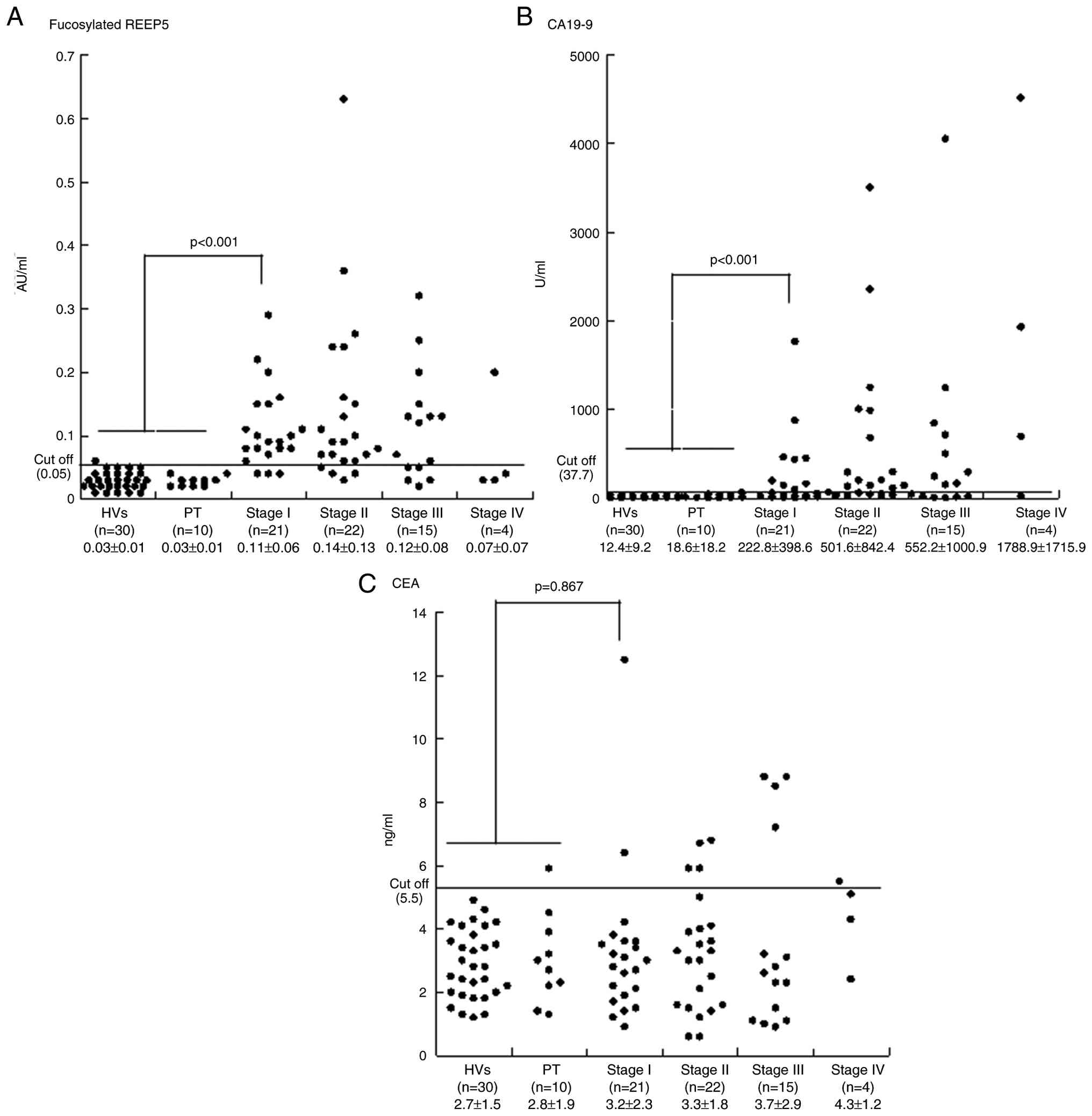
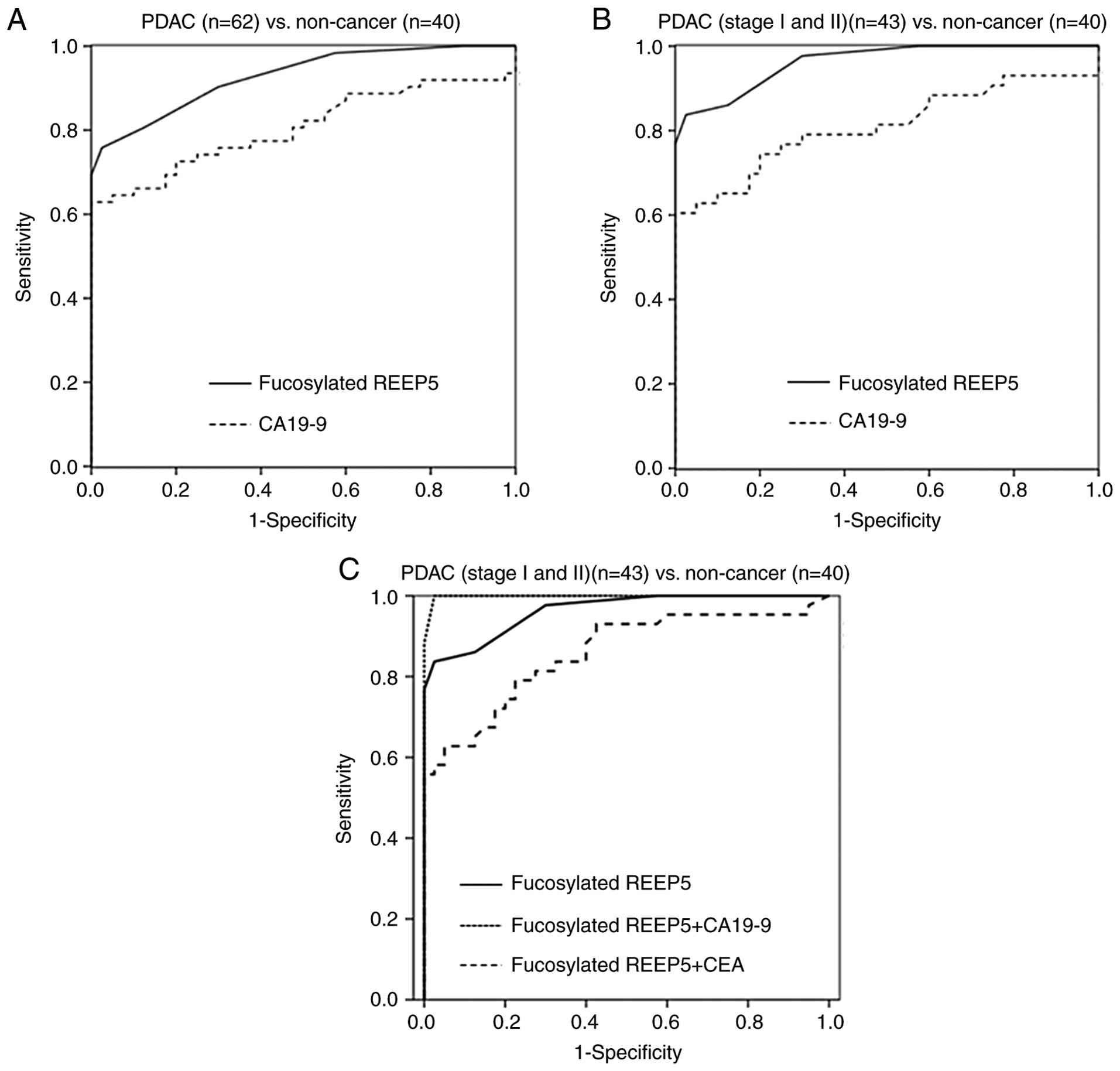
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49. 2024.
|
|
2
|
Hu ZI and O'Reilly EM: Therapeutic
developments in pancreatic cancer. Nat Rev Gastroenterol Hepatol.
21:7–24. 2024. View Article : Google Scholar
|
|
3
|
Hanada K, Shimizu A, Tsushima K, Ikeda M
and Tazuma S: Diagnosis of the early-stage pancreatic cancer and
collaboration of local clinics and hospitals. Nippon Rinsho.
81:513–518. 2023.
|
|
4
|
Kanno A, Masamune A, Hanada K, Maguchi H,
Shimizu Y, Ueki T, Hasebe O, Ohtsuka T, Nakamura M, Takenaka M, et
al: Multicenter study of early pancreatic cancer in Japan.
Pancreatology. 18:61–67. 2018. View Article : Google Scholar
|
|
5
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar
|
|
6
|
Welsh JA, Goberdhan DCI, O'Driscoll L,
Buzas EI, Blenkiron C, Bussolati B, Cai H, Di Vizio D, Driedonks
TAP, Erdbrügger U, et al: Minimal information for studies of
extracellular vesicles (MISEV2023): From basic to advanced
approaches. J Extracell Vesicles. 13:e124042024. View Article : Google Scholar
|
|
7
|
Koba T, Takeda Y, Narumi R, Shiromizu T,
Nojima Y, Ito M, Kuroyama M, Futami Y, Takimoto T, Matsuki T, et
al: Proteomics of serum extracellular vesicles identifies a novel
COPD biomarker, fibulin-3 from elastic fibres. ERJ Open Res.
7:00658–2020. 2021. View Article : Google Scholar
|
|
8
|
Shiromizu T, Kume H, Ishida M, Adachi J,
Kano M, Matsubara H and Tomonaga T: Quantitation of putative
colorectal cancer biomarker candidates in serum extracellular
vesicles by targeted proteomics. Sci Rep. 7:127822017. View Article : Google Scholar
|
|
9
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015. View Article : Google Scholar
|
|
10
|
Phinney DG and Pittenger MF: Concise
review: MSC-derived exosomes for cell-free therapy. Stem Cells.
35:851–858. 2017. View Article : Google Scholar
|
|
11
|
Kasahara K, Narumi R, Nagayama S, Masuda
K, Esaki T, Obama K, Tomonaga T, Sakai Y, Shimizu Y and Adachi J: A
large-scale targeted proteomics of plasma extracellular vesicles
shows utility for prognosis prediction subtyping in colorectal
cancer. Cancer Med. 12:7616–7626. 2023. View Article : Google Scholar
|
|
12
|
Bandu R, Oh JW and Kim KP: Mass
spectrometry-based proteome profiling of extracellular vesicles and
their roles in cancer biology. Exp Mol Med. 51:1–10. 2019.
View Article : Google Scholar
|
|
13
|
Kim H, Kim DW and Cho JY: Exploring the
key communicator role of exosomes in cancer microenvironment
through proteomics. Proteome Sci. 17:52019. View Article : Google Scholar
|
|
14
|
Muraoka S, Kume H, Watanabe S, Adachi J,
Kuwano M, Sato M, Kawasaki N, Kodera Y, Ishitobi M, Inaji H, et al:
Strategy for SRM-based verification of biomarker candidates
discovered by iTRAQ method in limited breast cancer tissue samples.
J Proteome Res. 11:4201–4210. 2012. View Article : Google Scholar
|
|
15
|
Narumi R, Murakami T, Kuga T, Adachi J,
Shiromizu T, Muraoka S, Kume H, Kodera Y, Matsumoto M, Nakayama K,
et al: A strategy for large-scale phosphoproteomics and SRM-based
validation of human breast cancer tissue samples. J Proteome Res.
11:5311–5322. 2012. View Article : Google Scholar
|
|
16
|
Kume H, Muraoka S, Kuga T, Adachi J,
Narumi R, Watanabe S, Kuwano M, Kodera Y, Matsushita K, Fukuoka J,
et al: Discovery of colorectal cancer biomarker candidates by
membrane proteomic analysis and subsequent verification using
selected reaction monitoring (SRM) and tissue microarray (TMA)
analysis. Mol Cell Proteomics. 13:1471–1484. 2014. View Article : Google Scholar
|
|
17
|
Sogawa K, Yamanaka S, Takano S, Sasaki K,
Miyahara Y, Furukawa K, Takayashiki T, Kuboki S, Takizawa H, Nomura
F and Ohtsuka M: Fucosylated C4b-binding protein α-chain, a novel
serum biomarker that predicts lymph node metastasis in pancreatic
ductal adenocarcinoma. Oncol Lett. 21:1272021. View Article : Google Scholar
|
|
18
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter
3. Unit 3.22. 2006.
|
|
19
|
Lamparski HG, Metha-Damani A, Yao JY,
Patel S, Hsu DH, Ruegg C and Le Pecq JB: Production and
characterization of clinical grade exosomes derived from dendritic
cells. J Immunol Methods. 270:211–226. 2002. View Article : Google Scholar
|
|
20
|
Muraoka S, Hirano M, Isoyama J, Nagayama
S, Tomonaga T and Adachi J: Comprehensive proteomic profiling of
plasma and serum phosphatidylserine-positive extracellular vesicles
reveals tissue-specific proteins. iScience. 25:1040122022.
View Article : Google Scholar
|
|
21
|
Masuda T, Tomita M and Ishihama Y: Phase
transfer surfactant-aided trypsin digestion for membrane proteome
analysis. J Proteome Res. 7:731–740. 2008. View Article : Google Scholar
|
|
22
|
Rappsilber J, Mann M and Ishihama Y:
Protocol for micro-purification, enrichment, pre-fractionation and
storage of peptides for proteomics using StageTips. Nat Protoc.
2:1896–1906. 2007. View Article : Google Scholar
|
|
23
|
Tyanova S, Temu T and Cox J: The MaxQuant
computational platform for mass spectrometry-based shotgun
proteomics. Nat Protoc. 11:2301–2319. 2016. View Article : Google Scholar
|
|
24
|
MacLean B, Tomazela DM, Shulman N,
Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC and
MacCosset MJ: Skyline: An open source document editor for creating
and analyzing targeted proteomics experiments. Bioinformatics.
26:966–968. 2010. View Article : Google Scholar
|
|
25
|
Narumi R, Shimizu Y, Ukai-Tadenuma M, Ode
KL, Kanda GN, Shinohara Y, Sato A, Matsumoto K and Ueda HR: Mass
spectrometry-based absolute quantification reveals rhythmic
variation of mouse circadian clock proteins. Proc Natl Acad Sci
USA. 113:E3461–E3467. 2016. View Article : Google Scholar
|
|
26
|
Miyahara Y, Takano S, Sogawa K, Tomizawa
S, Furukawa K, Takayashiki T, Kuboki S and Ohtsuka M: Prosaposin,
tumor-secreted protein, promotes pancreatic cancer progression by
decreasing tumor-infiltrating lymphocytes. Cancer Sci.
113:2548–2559. 2022. View Article : Google Scholar
|
|
27
|
Aoyagi Y, Isemura M, Suzuki Y, Sekine C,
Soga K, Ozaki T and Ichida F: Fucosylated alpha-fetoprotein as
marker of early hepatocellular carcinoma. Lancet. 2:1353–1354.
1985. View Article : Google Scholar
|
|
28
|
Food and Drug Administration, HHS Medical
devices; immunology and microbiology devices; classification of
AFP-L3% immunological test systems, . Final rule. Fed Regist.
70:57748–57750. 2005.
|
|
29
|
Szymendera JJ: Clinical usefulness of
three monoclonal antibody-defined tumor markers: CA 19-9, CA 50,
and CA 125. Tumour Biol. 7:333–342. 1986.
|
|
30
|
Konstantopoulos K and Thomas SN: Cancer
cells in transit: The vascular interactions of tumor cells. Annu
Rev Biomed Eng. 11:177–202. 2009. View Article : Google Scholar
|
|
31
|
Nakamori S, Kameyama M, Imaoka S, Furukawa
H, Ishikawa O, Sasaki Y, Kabuto T, Iwanaga T, Matsushita Y and
Irimura T: Increased expression of sialyl Lewisx antigen correlates
with poor survival in patients with colorectal carcinoma:
Clinicopathological and immunohistochemical study. Cancer Res.
53:3632–3637. 1993.
|
|
32
|
Park SH, Zhu PP, Parker RL and Blackstone
C: Hereditary spastic paraplegia proteins REEP1, spastin, and
atlastin-1 coordinate microtubule interactions with the tubular ER
network. J Clin Investig. 120:1097–1110. 2010. View Article : Google Scholar
|
|
33
|
Esteves T, Durr A, Mundwiller E, Loureiro
JL, Boutry M, Gonzalez MA, Gauthier J, El-Hachimi KH, Depienne C,
Muriel MP, et al: Loss of association of REEP2 with membranes leads
to hereditary spastic paraplegia. Am J Hum Genet. 94:268–277. 2014.
View Article : Google Scholar
|
|
34
|
Kumar D, Golchoubian B, Belevich I,
Jokitalo E and Schlaitz AL: REEP3 and REEP4 determine the tubular
morphology of the endoplasmic reticulum during mitosis. Mol Biol
Cell. 30:1377–1389. 2019. View Article : Google Scholar
|
|
35
|
Schlaitz AL, Thompson J, Wong CCL, Yates
JR III and Heald R: REEP3/4 ensure endoplasmic reticulum clearance
from metaphase chromatin and proper nuclear envelope architecture.
Dev Cell. 26:315–323. 2013. View Article : Google Scholar
|
|
36
|
Behrens M, Bartelt J, Reichling C, Winnig
M, Kuhn C and Meyerhof W: Members of RTP and REEP gene families
influence functional bitter taste receptor expression. J Biol Chem.
281:20650–20659. 2006. View Article : Google Scholar
|
|
37
|
Liu GH, Tan XY, Xu ZY, Li JX, Zhong GH,
Zhai JW and Li MY: REEP3 is a potential diagnostic and prognostic
biomarker correlated with immune infiltration in pancreatic cancer.
Sci Rep. 14:138342024. View Article : Google Scholar
|
|
38
|
Yao L, Xie D, Geng L, Shi D, Huang J, Wu
Y, Lv F, Liang D, Li L, Liu Y, et al: REEP5 (receptor accessory
protein 5) acts as a sarcoplasmic reticulum membrane sculptor to
modulate cardiac function. J Am Heart Assoc. 7:e0072052018.
View Article : Google Scholar
|
|
39
|
Li M, Rao M, Chen K, Zhou J and Song J:
Selection of reference genes for gene expression studies in heart
failure for left and right ventricles. Gene. 620:30–35. 2017.
View Article : Google Scholar
|
|
40
|
Chiamvimonvat N and Song LS: LRRC10
(leucine-rich repeat containing protein 10) and REEP5 (receptor
accessory protein 5) as novel regulators of cardiac
excitation-contraction coupling structure and function. J Am Heart
Assoc. 7:e0082602018. View Article : Google Scholar
|
|
41
|
Park CR, You DJ, Park S, Mander S, Jang
DE, Yeom SC, Oh SH, Ahn C, Lee SH, Seong JY and Hwang JI: The
accessory proteins REEP5 and REEP6 refine CXCR1-mediated cellular
responses and lung cancer progression. Sci Rep. 6:390412016.
View Article : Google Scholar
|
|
42
|
Tian Z, He W, Tang J, Liao X, Yang Q, Wu Y
and Wu G: Identification of important modules and biomarkers in
breast cancer based on WGCNA. Onco Targets Ther. 13:6805–6817.
2020. View Article : Google Scholar
|
|
43
|
Kimura K, Ohto M, Saisho H, Unozawa T,
Tsuchiya Y, Morita M, Ebara M, Matsutani S and Okuda K: Association
of gallbladder carcinoma and anomalous pancreaticobiliary ductal
union. Gastroenterology. 89:1258–1265. 1985. View Article : Google Scholar
|
|
44
|
Athanasiou A, Kureshi N, Wittig A, Sterner
M, Huber R, Palma NA, King T and Schiess R: Biomarker discovery for
early detection of pancreatic ductal adenocarcinoma (PDAC) using
multiplex proteomics technology. J Proteome Res. 24:315–322. 2025.
View Article : Google Scholar
|
|
45
|
Tada K, Ohta M, Hidano S, Watanabe K,
Hirashita T, Oshima Y, Fujnaga A, Nakanuma H, Masuda T, Endo Y, et
al: Fucosyltransferase 8 plays a crucial role in the invasion and
metastasis of pancreatic ductal adenocarcinoma. Surg Today.
50:767–777. 2020. View Article : Google Scholar
|
|
46
|
Pinho SS and Reis CA: Glycosylation in
cancer: Mechanisms and clinical implications. Nat Rev Cancer.
15:540–555. 2015. View Article : Google Scholar
|
|
47
|
Watanabe K, Ohta M, Yada K, Komori Y,
Iwashita Y, Kashima K and Inomata M: Fucosylation is associated
with the malignant transformation of intraductal papillary mucinous
neoplasms: A lectin microarray-based study. Surg Today.
46:1217–1223. 2016. View Article : Google Scholar
|