High expression of eIF4A1 promotes angiogenesis through the NF‑κB/VEGFA pathway and predicts poor prognosis in gastric cancer
- Authors:
- Published online on: July 17, 2025 https://doi.org/10.3892/or.2025.8951
- Article Number: 118
-
Copyright: © Zhu et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Gastric cancer (GC) is the fourth most common cancer and the second leading cause of cancer-associated deaths worldwide (1). After lung and liver cancers, GC constitutes 20% of the total global burden of disability-adjusted life-years among men, highlighting its notable impact on global public health (2). Approximately 37% of GC cases occur in China, with 358, 000 new cases each year (3). The 5-year survival rate of patients with GC is unsatisfactory, since >80% of patients with GC are diagnosed at an advanced stage (4). Therefore, it is necessary to identify novel biomarkers for GC diagnosis and treatment.
Eukaryotic translation initiation factor 4A (eIF4A, also known as DDX2) is the most well-characterized helicase required for translation initiation, and serves a key role in unwinding secondary structures in the 5′UTR of mRNA (5). Translation initiation is facilitated by the eIF heterotrimeric complex, including the cap-binding protein eIF4E, the scaffold protein eIF4G and eIF4A, which is an ATP-dependent DEAD box RNA helicase (6). Both eIf4E and eIf4G are activated by the eIF4A protein, which has three isoforms; however, only two mammalian isoforms are involved in translation, namely eIF4A1 (DDX2A) and eIF4A1I (DDX2B). Furthermore, eIF4A1 is the preferred binding partner of the eIF4F complex compared with eIF4A1I, and is essential for cell proliferation (7,8). In addition, eIF4A1 is expressed at higher levels in proliferating cells, whereas eIF4A1I is upregulated in cells with a low proliferative capacity (9).
The dysregulation of mRNA translation is an important step in cancer progression, and translation initiation is the main rate-limiting step in protein expression (10). Furthermore, eIF4A1 is an important helicase that is involved in virtually all aspects of RNA helicase activities. Certain proteins, the activity of which is modulated by eIF4A1, show expression alterations in numerous types of cancer. eIF4B is upregulated in diffuse large B-cell lymphoma and is associated with poor prognosis (11). eIF4A promotes T-cell acute lymphoblastic development in vivo and is required for leukemia maintenance (12). Additionally, a small molecule inhibitor of the translation factor eIF4A, silvestrol, has been shown to reduce MYC translation and to inhibit tumor growth in a mouse model of colorectal tumorigenesis (13). Numerous natural molecules targeting eIF4A1 have a marked effect on tumor suppression, such as hippuristanol (14). However, the role of eIF4A1 in the angiogenesis of GC cells remains unknown; therefore, the present study aimed to determine the influence of eIF4A1 expression on patient prognosis and angiogenesis in GC.
Materials and methods
EIF4A1 mRNA in GC tissues
The RNA-seq data of 375 GC tissues and 32 normal adjacent tissues were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), and analyzed using the UALCAN website (http://ualcan.path.uab.edu) and the Home For Researchers (https://www.home-for-researchers.com) using Wilcoxon rank sum test and Kruskal-Wallis test with Dunn's post hoc test.
Patient tissue specimens and clinical information collection
A total of 732 samples, including 43 chronic gastritis tissues, 56 intestinal metaplasia tissues, 29 low-grade intraepithelial neoplasia tissues, 26 high-grade intraepithelial neoplasia tissues, 446 carcinoma tissues and 132 pericarcinomatous tissues from patients who donated carcinoma tissues, were collected from the Department of Clinical Pathology, Nantong University Hospital (Nantong, China) between April 2003 and October 2010. In addition, 1,026 samples, including 35 chronic gastritis tissues, 31 intestinal metaplasia tissues, 35 low-grade intraepithelial neoplasia tissues, 28 high-grade intraepithelial neoplasia tissues, 737 carcinoma tissues and 160 pericarcinomatous tissues, were collected from the Nanjing First Hospital (Nanjing, China) between July 2008 and May 2012. These patients included 582 men and 884 women, with a median age of 49 years (range, 26–83 years). None of the patients received radiation therapy, chemotherapy or immunotherapy before surgery or biopsy for pathological examination. The clinicopathological information of the patients included sex, age, histological type, differentiation, tumor invasion, lymph node metastases, distant metastasis, and preoperative CEA and Ca 19-9 levels. The pathological diagnosis was made according to the World Health Organization (WHO) Collaborating Centre for Gastric Cancer. The present study was approved by the Ethics Committee of Nanjing Medical University [(2017) approval no. 335; Nanjing, China]. Written informed consent was obtained from all patients enrolled in the study.
Tissue microarrays (TMAs) and immunohistochemistry (IHC) analysis
Every microarray included 7×10 points and was generated using the Tissue Microarray System (cat. no. UT06; Quick-Ray; UNITMA Co., Ltd.). The fresh tissues were fixed in 10% neutral formalin at room temperature for 24 h, and then embedded in paraffin. Every point (diameter, 2 mm) was taken from independent paraffin-embedded tissues prepared earlier. A total of 30 TMAs were prepared and cut into 4-µm sections. The tissue sections were deparaffinized and rehydrated in xylene and a descending series of ethanol concentrations, and 3% hydrogen peroxide (100°C, 20 min) was used to inhibit endogenous peroxidase (15). Then, antigen retrieval was performed using sodium citrate acid buffer (pH, 6.0). The sections were then incubated with anti-eIF4A1 antibody (1:200; cat. no. ab31217; Abcam) overnight at 4°C. The Envision peroxidase kit (Dako; Agilent Technologies, Inc.) was used for to incubate the sections at room temperature for 1 h, and they were then incubated with 3,3′-diaminobenzidine solution for coloration and were counterstained with hematoxylin at room temperature for 10 min, respectively. The primary antibody was replaced with PBS as a negative control (NC) (14). Gastritis tissues and intestinal metaplasia tissues were treated as internal controls. eIF4A1 protein expression was evaluated by Vectra 3.0 (PerkinElmer, Inc.) using the ‘Automatically Identify Sample Points’ function. The staining intensity was defined as follows: 0, no staining; 1, weak staining; 2, moderate staining; and 3, intense staining. The final score was calculated by multiplying the staining intensity by the percentage of stained cells/total cells, as follows IHC score=(3 × percentage of strongly stained cells + 2 × percentage of moderately stained cells + 1 × percentage of weakly stained cells) ×100. The minimum score was 0 (no staining) and the maximum score was 300 (3×100%).
Cell culture
Human GC cell lines FU97 and HGC27, and the human umbilical vein endothelial cell (HUVEC) line were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences, and preserved in the National Health Commission Key Laboratory of Antibody Technique (Nanjing Medical University). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin. A constant environment at 37°C and 5% CO2 was utilized to maintain all cells.
Construction of stable cell lines
Lentiviruses containing eIF4A1 cDNA (pLV3-CMV-eIF4A1-CopGFP-Puro) or short hairpin (sh)RNA against eIF4A1 (pLV3-U6-eIF4A1-shRNA-CopGFP-Puro) were produced by GeneCopoeia, Inc, as well as their controls (NC and shNC). The target sequence of the shRNA was 5′-GCCGTAAAGGTGTGGCTATTA-3′, and the shNC sequence was 5′-GTTCTCCGAACGTGTCACGTT-3′. For eIF4A1 overexpression, the NC was an empty plasmid vector without the target gene. FU97 cells infected with the lentivirus containing eIF4A1 cDNA, whereas HGC27 cells were transfected with the lentivirus encoding eIF4A1 shRNA. GC cells in each group were seeded in 96-well plates at a density of 1×104 cells/well. After 24 h, the original medium was replaced with serum-free medium and an appropriate amount of lentivirus was added, according to the following formula: Virus volume=(MOI × number of cells)/virus titer (MOI=10). After incubation with lentiviruses for 8 h, the GC cells were cultured in complete medium for 48 h. Cells were cultured with puromycin dihydrochloride (2 µg/ml, cat. no. A1113803; Gibco; Thermo Fisher Scientific, Inc.) for 2 weeks.
Conditioned medium (CM)
GC cells were routinely cultured for 48 h and then incubated with fresh DMEM for 24 h. The supernatant was subsequently filtered through 0.22-µm filters and stored at −80°C until required.
Western blotting
Total protein from each group of GC cells was extracted using RIPA lysis buffer (cat. no. 89901; Thermo Fisher Scientific, Inc.) and protein concentration was detected using the BCA kit (cat. no. P0012S; Beyotime Institute of Biotechnology). The total protein was mixed with sample loading buffer in equal proportion, heated at 95°C for 10 min and stored at −20°C. The protein samples (10 µg/lane) were separated by SDS-PAGE on 10% polyacrylamide gels and transferred to a PVDF membrane. The membrane was blocked with 5% skim milk at room temperature for 1 h, then incubated with anti-eIF4A1 antibody (1:1,000; cat. no. ab31217; Abcam), VEGFA (1:1,000; cat. no. MA00045; Boster Biological Technology), NF-κB (1:1,000; cat. no. ab32536; Abcam) and GAPDH (1:1,000; cat. no. AF2823; Beyotime Institute of Biotechnology) at 4°C overnight, and then with a HRP-labeled sheep anti-rabbit IgG antibody (1:2,000; cat. no. ab6721; Abcam) at room temperature for 1 h. For blot visualization, the SuperSignal West Pico PLUS chemiluminescence substrate (cat. no. 34578; Thermo Fisher Scientific, Inc.) was applied to cover the membrane surface, and the ChemiDoc XRS+ system and Image Lab 5.1 (Bio-Rad Laboratories, Inc.) were used.
Cell counting kit 8 (CCK-8) assay
HUVECs were plated in 96-well plates at a density of 1×104/well with CM. The original culture medium was removed at 12, 24, 36 and 48 h. Subsequently, 10 µl CCK-8 reagent (cat. no. CD04; Dojindo Laboratories, Inc.) and 90 µl culture medium were added to each well, and incubated at 37°C for 3 h without light. The absorbance value of each well was detected at 450 nm using a microplate reader.
Wound healing assay
HUVECs were plated in 6-well plates at a density of 5×105 cells/well. After adherence, the medium was replaced with serum-free CM and the cells were cultured overnight. A 100-µl pipette tip was utilized to scratch the cell surface when cell confluence approached 90%. After washing, the culture was continued with serum-free CM. Images of the cell culture plates were captured at 0 and 24 h with a light microscope (IX70; Olympus Corporation), and were analyzed using Image Pro Plus 6.0 software (Media Cybernetics, Inc.). Three scratch fields were randomly selected for each group to measure the scratch distance.
Transwell assay
HUVECs were plated in a Transwell chamber (pore size, 8 µm; cat. no. 3422; Corning, Inc.) at a density of 5×104/well with 200 µl serum-free CM. The chambers were placed in 12-well plates, and 900 µl CM containing 20% FBS was added to the lower wells. The upper surface of the chamber membrane was gently wiped with cotton swabs after 24 h, and the cells on the other surface were then fixed with methanol for 30 min and stained with 0.1% crystal violet solution for 30 min at room temperature. The number of cells on the membrane was counted and images were captured with a microscope (cat. no. IX70; OLYMPUS).
Angiogenesis assay
Matrigel was thawed and diluted at 4°C to cover the bottom of a 96-well plate, which was placed in an incubator at 37°C for 30 min. HUVECs were then plated in the 96-well plates at a density of 2×104/well and the formation of the lumen was observed under a light microscope (IX70; Olympus Corporation) after 8 h. Five fields were randomly selected for assessment and images were collected. The results were analyzed using ImageJ 1.5.4 software (National Institutes of Health).
Subcutaneous tumor model in nude mice
A total of 12 BALB/c nude mice (male; weight, 18–22 g; age, 6 weeks) were purchased from Charles River Laboratories, Inc. Mice were housed at a temperature of 20–26°C and a humidity of 40–70%, were provided free access to sterilized rodent chow and sterilized drinking water, and were maintained under a fixed 12-h light/dark cycle. The HGC27-shNC and HGC27-sheIF4A1 cells were used to construct the models. For each group (n=6 mice), cells in the logarithmic growth phase were prepared as suspensions with a density of 1×107/ml, which were then injected into the axilla of the upper limb of nude mice (100 µl/mouse). After 5 weeks, the nude mice were intravenously injected with sodium pentobarbital (50 mg/kg) and were sacrificed by cervical dislocation. Subcutaneous tumors were then separated and fixed in 10% formalin at room temperature for 24 h before IHC staining.
Bioinformatics analysis of the relationship between eIF4A1 and components of the tumor microenvironment (TME)
The UALCAN platform and the Home for Researcher platform were used for analysis of the eIF4A1 expression, and its correlation with CD31, VEGFA, NF-κB1 and NF-κB2 based on TCGA database (https://portal.gdc.cancer.gov/) using Pearson correlation coefficient. TCGA datasets or Gene Expression Omnibus (GEO) datasets (https://www.ncbi.nlm.nih.gov/geo/) were downloaded and analyzed using the R software package (4.3.3) (https://cran.r-project.org/). In detail, the ImmuCellAI algorithm (https://guolab.wchscu.cn/ImmuCellAI/#!/analysis#abundance_result) was applied to the gene expression profiles of the GEO datasets (GSE62254, GSE15459 and GSE84426) to estimate the abundance scores of 24 immune cell types for each sample (16–18). The dataset was divided into high and low expression groups based on the median expression level of the eIF4A1 gene. Differences in immune cell abundance between the two groups were assessed using the Wilcoxon rank-sum test. Boxplots were generated with the ggplot2 package (https://cran.r-project.org/web/packages/ggplot2/) to visualize the correlation between eIF4A1 expression and components of the TME. Multiple regression analysis was used to assess the association between eIF4A1 expression and VEGFA.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from FU97 and HGC27 cells using TRIzol reagent (cat. no. 15596026CN; Invitrogen; Thermo Fisher Scientific, Inc.) and was then reverse transcribed into cDNA using the Maxima H Minus First Strand cDNA Synthesis Kit (cat. no. K1652; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. qPCR was performed using the TB Green Premix Ex Taq (cat. no. RR420Q; Takara Bio, Inc.) on the StepOne Real-Time PCR System. The amplification conditions were as follows: 95°C for 10 min; followed by 40 cycles at 95°C for 15 sec and 56°C for 1 min; and finally, 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. Primer synthesis was completed by Shanghai Shenggong Biology Engineering Technology Service, Ltd., with the following sequences: VEGFA, forward 5′-AGGGCAGAATCATCACGAAGT-3′, reverse 5′- AGGGTCTCGATTGGATGGCA-3′; VEGFB, forward 5′-GAGATGTCCCTGGAAGAACACA-3′, reverse 5′-GAGTGGGATGGGTGATGTCAG-3′; VEGFC, forward 5′-GAGGAGCAGTTACGGTCTGTG-3′, reverse 5′-TCCTTTCCTTAGCTGACACTTGT-3′; VEGFD, forward 5′-TCCCATCGGTCCACTAGGTTT-3′, reverse 5′-AGGGCTGCACTGAGTTCTTTG-3′; and GAPDH, forward 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′. GAPDH was used as the reference gene and the relative expression levels of target genes were calculated using the 2−ΔΔCq method (19).
Statistical analysis
SPSS software version 18.0 (SPSS, Inc.) was used to analyze data in the present study. X-tile 3.6.1 software (The Rimm Lab at Yale University; http://medicine.yale.edu/lab/rimm/research/software/) was used to calculate the cut-off value for the high and low eIF4A1 groups. The measurement data are presented as the mean ± standard deviation. The Pearson χ2 test was used to determine the association between eIF4A1 and clinicopathological features. Cumulative survival was analyzed using the Kaplan-Meier method, and the log-rank test was used to obtain P-values. The Cox proportional risk regression model was established to analyze univariate and multivariate factors. All in vitro experiments were repeated three times. An independent t-test was used to compare the differences between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
eIF4A1 is upregulated in GC
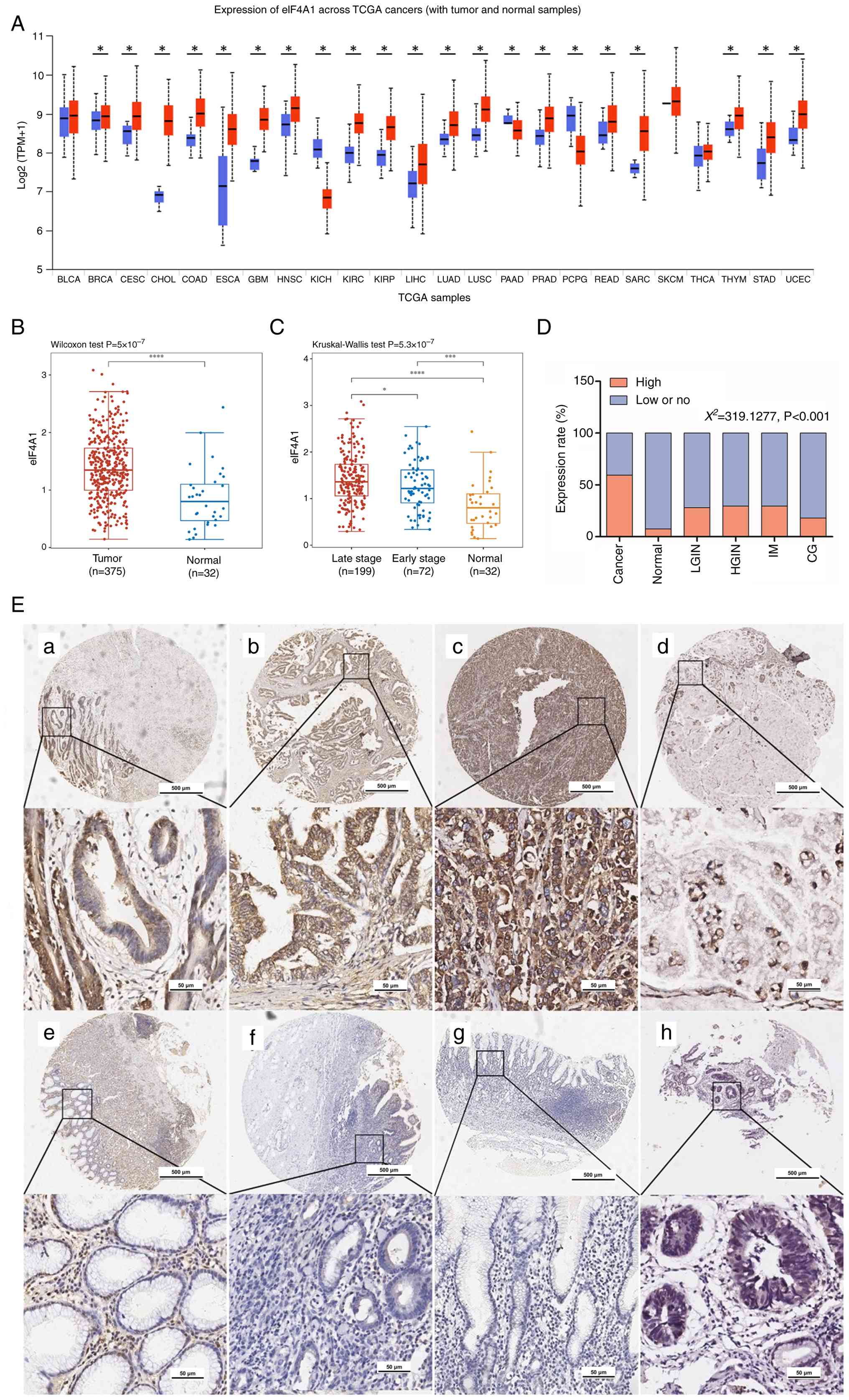
To explore the expression of eIF4A1 in GC, data from TCGA database were first analyzed. According to pan-cancer analysis using the UALCAN website, eIF4A1 mRNA expression levels were increased in GC (stomach adenocarcinoma, STAD) compared with those in normal gastric tissues from patients with GC (Fig. 1A). Moreover, the mRNA expression levels of eIF4A1 were significantly upregulated in 375 tumor tissues compared with in 32 peripheral normal tissues from patients with GC, based on analysis of TCGA-STAD datasets (Fig. 1B). Furthermore, eIF4A1 mRNA expression levels in patients with late-stage GC were higher than those in patients with early-stage (Fig. 1C).
Protein expression was evaluated by IHC in tissues from patients with GC, and the results indicated that eIF4A1 protein was mainly expressed in the cytoplasm (Fig. 1E). The cut-off value of staining scores was determined as 110 using X-tile 3.6.1 software, which maximized the survival difference between the two groups of data after grouping. Scores 0–110 were regarded as low or no eIF4A1 expression, whereas those 111–300 were regarded as high eIF4A1 expression. The results indicated that eIF4A1 was relatively highly expressed in carcinoma, at 59.26% (701/1,183). However, high eIF4A1 expression was found in only 14.10% (11/78) of patients with chronic gastritis and 7.19% (21/292) of the pericarcinomatous tissue. Low-grade and high-grade intraepithelial neoplasia showed a slightly higher percentage of high eIF4A1 protein expression than pericarcinomatous tissues, at 26.56% (17/64) and 29.63% (16/54), respectively (Fig. 1D).
eIF4A1 protein expression is associated with clinicopathological factors in patients with GC
The association between clinicopathological features and eIF4A1 expression in the TMA samples was subsequently assessed (Table I). EIF4A1 expression in GC was associated with age (χ2=19.9945, P<0.001), differentiation (χ2=8.4049, P=0.015), depth of invasion (χ2=14.3348, P=0.006), distant metastasis (χ2=20.5599, P<0.001) and Tumor-Node-Metastasis (TNM) stage (χ2=33.2548, P<0.001). However, eIF4A1 high expression was not significantly associated with sex, histological type, lymph node metastasis, and preoperative CEA and CA 19-9 (P>0.05).
Table I.eIF4A1 protein expression level and the clinicopathological characteristics of patients with gastric cancer. |
eIF4A1 expression predicts a poor prognosis for patients with GC
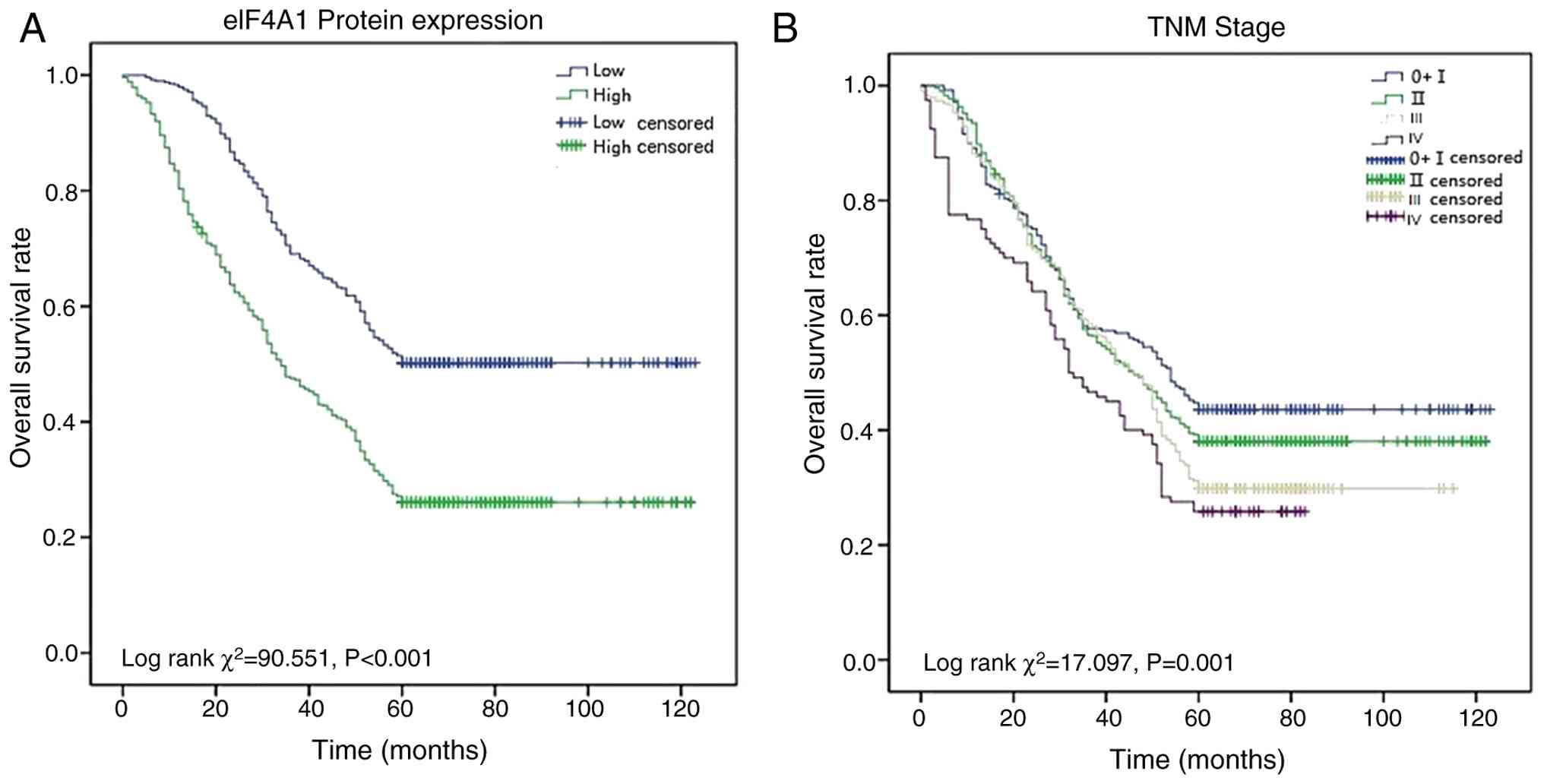
Univariate and multivariate Cox proportional hazards analyses were performed to investigate the parameters that might affect the overall survival (OS) of patients with GC (Table II). In the univariate analysis, the OS of patients with GC was associated with eIF4A1 expression (HR=2.060, P<0.001), age (HR=1.286, P=0.001), distant metastasis (HR=4.473, P<0.001), TNM stage (HR=1.175, P<0.001) and differentiation (HR=1.189, P<0.001). Subsequently, the significant factors in the univariate analysis underwent multivariate analysis, and eIF4A1 was revealed to be an independent prognostic parameter for patients with GC (HR=1.174, P<0.001). The Kaplan-Meier survival curves revealed that advanced TNM stage and high eIF4A1 were associated with poor OS in patients with GC (P<0.001; Fig. 2).
Table II.Univariate and multivariate analyses of the prognostic factors for overall survival in gastric cancer. |
eIF4A1 enhances the proangiogenic activity of GC cells
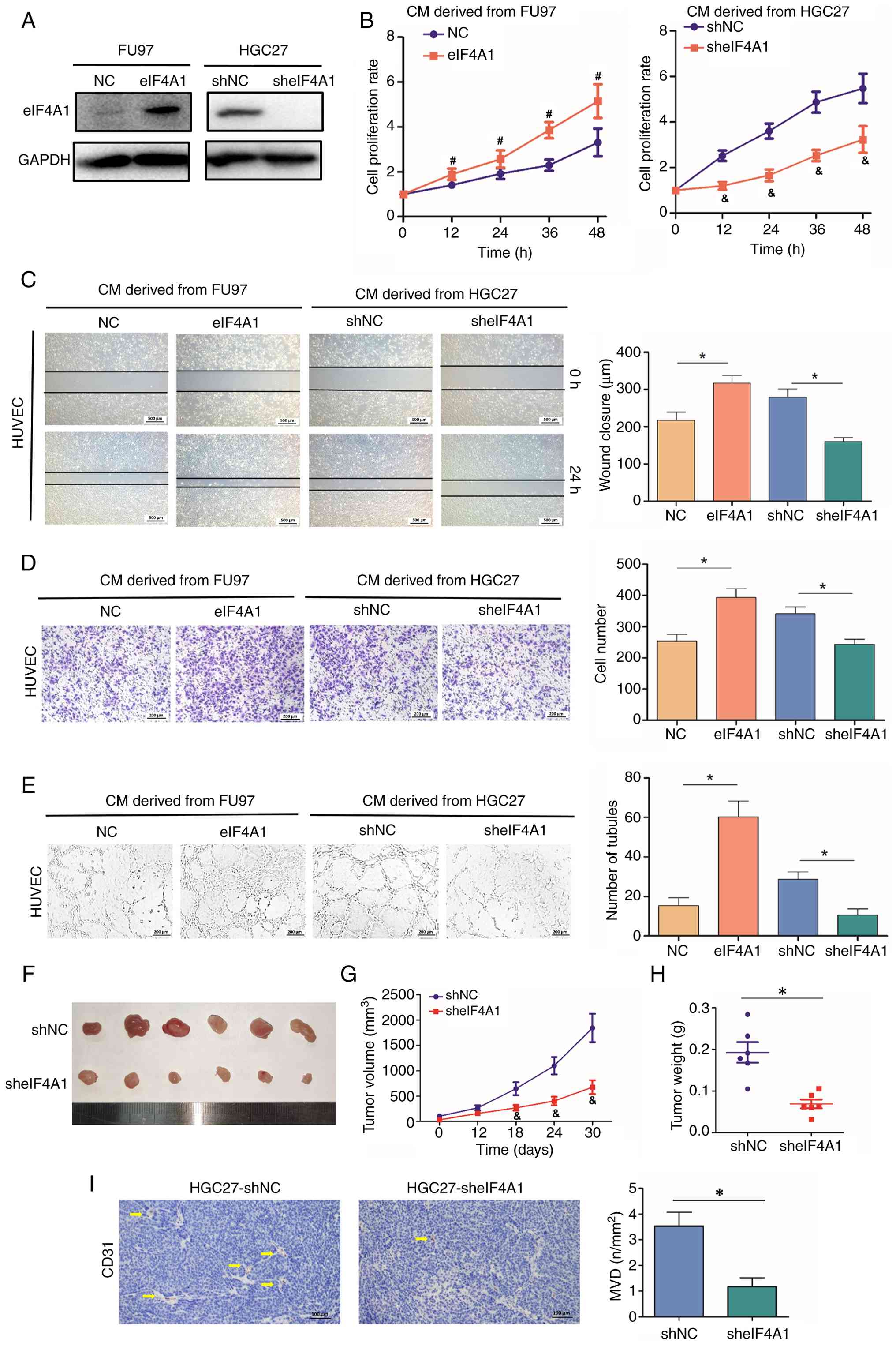
To validate the influence of eIF4A1 on the angiogenesis of GC cells, eIF4A1 was overexpressed in FU97 cells and knocked down in HGC27 cells using lentiviral vectors. Western blotting confirmed the alterations in eIF4A1 expression (P<0.05; Fig. 3A). Subsequently, HUVECs were treated with the CM of GC cells to explore the proangiogenic role of eIF4A1. eIF4A1 exhibited a significant positive effect on HUVEC proliferation (P<0.05; Fig. 3B). Furthermore, the wound healing and Transwell assays demonstrated the effects of eIF4A1 on the promotion of HUVEC migration (P<0.05; Fig. 3C and D). CM derived from eIF4A1 overexpression cells also significantly promoted HUVEC tube formation, whereas CM derived from eIF4A1 knockdown cells had the opposite effect (P<0.05; Fig. 3E). The present study also constructed a subcutaneous tumor model in nude mice using HGC27-sheIF4A1 cells; the results demonstrated that eIF4A1 knockdown inhibited tumor growth (P<0.05; Fig. 3F-H), which was the same as the findings of a previous study (20). Therefore, the current study assessed the influence of eIF4A1 on tumor angiogenesis. The subcutaneous xenograft tumor model demonstrated that HGC27-sheIF4A1 induced fewer blood vessels in tumor tissues than HGC27-shNC (P<0.05; Fig. 3I).
eIF4A1 promotes VEGFA through the NF-κB pathway
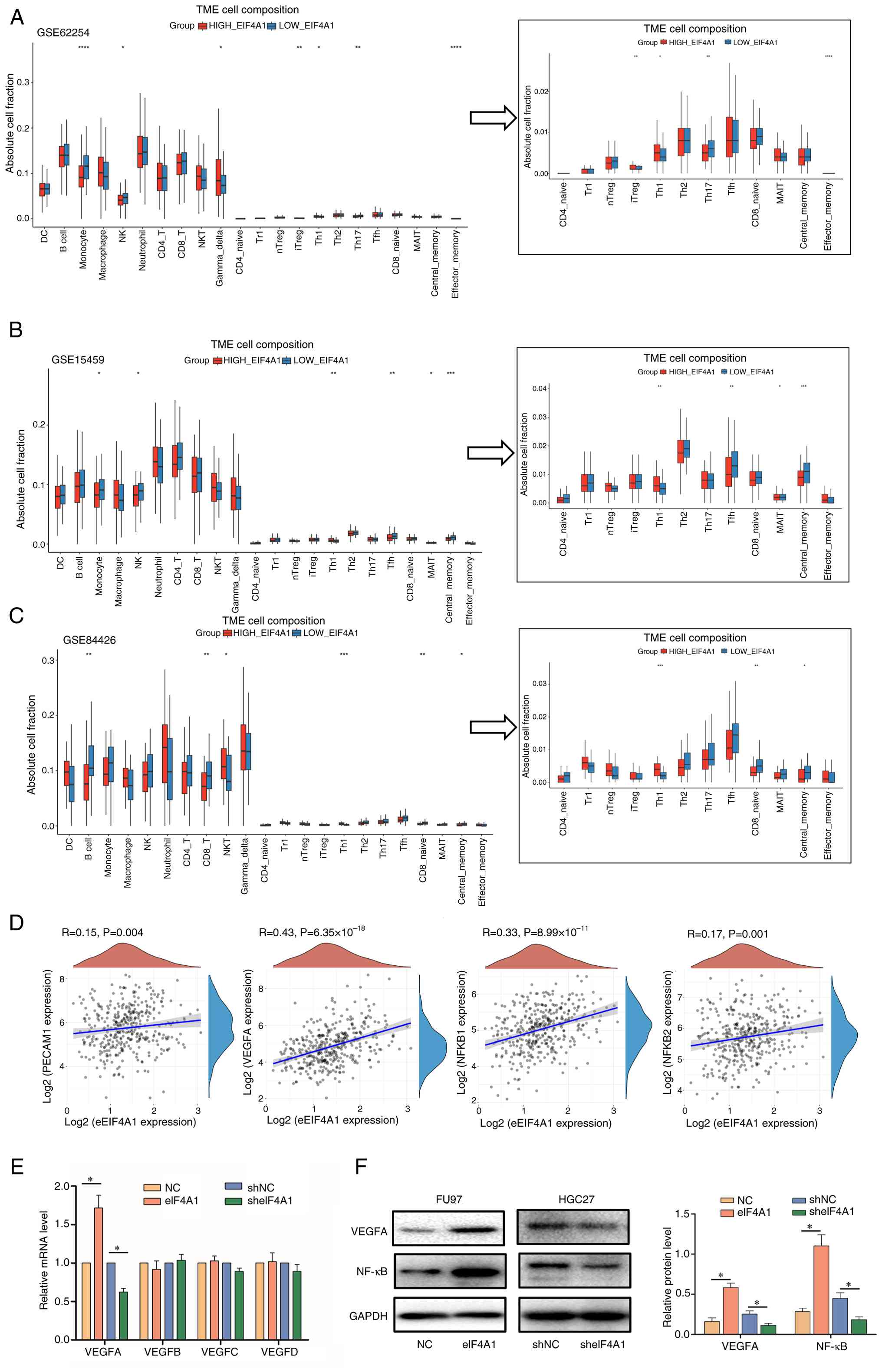
The present study assessed the relationship between eIF4A1 expression and components of the TME. The results showed that the elevated expression of eIF4A1 was associated with decreased monocytes, natural killer (NK) cells, macrophages, CD8+ T cells and B cells, but was associated with increased regulatory T (Treg) and T helper 2 (Th2) cells according to the three datasets (P<0.05; Fig. 4A-C). Subsequently, the present study evaluated the correlation between eIF4A1 mRNA expression and angiogenesis. It was revealed that increased eIF4A1 expression was significantly positively correlated with the expression levels of, VEGFA, NF-κB1, NF-κB2 and CD31 (also called platelet endothelial cell adhesion molecule 1, a marker of endothelial cells); however the correlation with the latter two genes was relatively weak (P<0.05; Fig. 4D). VEGF is a vital factor regulating cancer angiogenesis in the TME (21). The present study subsequently detected the effects of eIF4A1 dysregulation on the VEGF family, and it was revealed that eIF4A1 overexpression could promote the mRNA expression levels of VEGFA, whereas knockdown of eIF4A1 exhibited the opposite trend (P<0.05; Fig. 4E). Subsequently, the associated of eIF4A1 with VEGFA was confirmed at the protein level by western blotting (P<0.05; Fig. 4F). Meanwhile, eIF4A1 overexpression promoted NF-κB expression, whereas eIF4A1 knockdown decreased the levels of NF-κB in GC cells (P<0.05; Fig. 4F).
Discussion
As a key member of the DEAD-box proteins, eIF4A has two RecA-like domains, which contain the core motifs necessary for ATP binding and hydrolysis, and RNA binding and melting (22). The eIF4A1 protein is best characterized as an important RNA helicase (23). Since RNA unwinding and ribosome binding are crucial steps in transcriptional initiation, which regulates the rate-limiting step (24), high eIF4A1 expression markedly increases the speed of protein transcription in GC. Furthermore, the high expression of eIF4A1 protein signifies a substantial demand for RNA and proteins within the cell, a scenario often triggered by cellular injury or abnormal proliferative responses. In the present study, high eIF4A1 expression was detected in only 7.19% of pericarcinomatous normal tissues, which may be attributed to cell division or other normal metabolic processes, and in 14.10% of chronic gastritis tissues, which may be associated with cell injury and repair. Notably, it was detected in >25% of intestinal metaplasia, and low or high-grade intraepithelial neoplasia cases, and in 59.26% of carcinoma cases, which indicated abnormal tumor metabolism and a higher protein transcription demand. eIF4A1, in the malignant phenotype (such as uncontrolled proliferation, anti-apoptosis and metastasis), induces the expression of numerous oncogenes, including cyclin D1, Bcl-x and MUC1 (12,14,25), and inhibition of eIF4A1 with silvestrol has been shown to improve chemosensitivity in mouse models of lymphoma (26). Thus, eIF4A1 upregulation may selectively increase oncogene protein expression and abnormal metabolism in cells that are prepared for cancer cell proliferation and other activations.
The results of the present study suggested that high eIF4A1 protein was closely related to poor tumor differentiation, deep invasion and distant metastasis. Although Gao et al (20) also explored the associations between eIF4A1 levels and the clinicopathological features of 191 patients with GC, they only classified GC as intestinal-type, diffuse-type and mixture-type. It might be helpful for the investigation of the influence of eIF4A1 on GC if more types of samples were examined. In the present study, a total of 1,183 GC cases were separated into five types: Tubular, mucinous, mixed (tubular and mucinous), Signet ring cell and other, based on WHO classification guidelines (27). Notably, eIF4A1 expression was revealed to be upregulated in all types of GC tissues compared with adjacent gastric tissues. In addition, the use of multicenter large-scale sample data in the present study could further ensure the accuracy of the statistics. Furthermore, univariate and multivariate analyses indicated that eIF4A1 may be considered an independent prognostic biomarker in GC.
The TME refers to the environment in which tumor and tumor stem cells live, including tumor cells, stromal cells, surrounding immune cells and fibroblasts. The various immune cells in the TME can be tumor suppressors or tumor supporters. NK cells are vital fighters against tumor cells and their decreased infiltration predicts a poor outcome in GC (28). CD8+ T cells can specifically recognize and kill tumor cells, thus serving a key role in the antitumor immune response (29). B cells can not only differentiate into plasma cells and secrete antibodies, but also promote T cell-induced antitumor immunity through antigen presentation (30). Treg cells inhibits the immune response by expressing CTLA4, and secreting IL-10 and TGF-β, which promotes immune escape of tumor cells (31). Th2 cells antagonize the antitumor immune response of Th1 cells and are positively associated with tumor malignancy (32). The current study confirmed that high eIF4A1 was associated with decreased monocytes, NK cells, macrophages, CD8+ T cells and B cells, but was linked to increased Treg and Th2 cells, thus indicating that eIF4A1 might induce a suppressive immune microenvironment in GC.
Angiogenesis has a key role in the proliferation, invasion, migration and metastasis of GC (33). Tumor blood vessels not only provide oxygen, proteins and other nutrients for the metabolism of new tumor cells, but also promote the metastasis and settlement of tumor cells from the primary site to distant sites, resulting in tumor deterioration (34). Tumor cells can regulate the vascular environment, and proliferation and migration of endothelial cells through paracrine effects. The direct effect of eIF4A1 on the proliferation, apoptosis, migration and invasion of GC cells has been confirmed in several studies (26,35). However, little is currently known about the influence of eIF4A1 on GC cell angiogenesis. The present study confirmed that overexpression of eIF4A1 promoted the proliferation, migration and tube formation of HUVECs, whereas eIF4A1 knockdown inhibited these biological behaviors. In addition, a subcutaneous xenograft tumor constructed using HGC27-sheIF4A1 cells exhibited fewer microvessels than that in the control group. These results indicated that eIF4A1 could facilitate the angiogenesis of GC cells, which may further accelerate the proliferation and metastasis of GC.
VEGF is considered to be the most potent and highly specific regulator of angiogenesis (36). Upon binding to VEGFR1/2 on vascular endothelial cells, VEGF directly drives cell proliferation through activation of the PI3K-AKT and RAS-MAPK signaling cascades. Concomitantly, it induces endothelial cells to secrete additional angiogenic factors (such as PDGF) in a paracrine manner, thereby synergistically augmenting the angiogenic response (37). VEGF concentration has been shown to be significantly increased in GC, where it can accelerate the production of new blood vessels (38). The present study confirmed that eIF4A1 could promote the expression of VEGFA, a member of the VEGF family, thus suggesting that eIF4A1 promotes GC angiogenesis through increasing VEGFA secretion. Furthermore, NF-κB is a pleiotropic nuclear transcription factor that induces VEGFA expression and promotes angiogenesis (39–41). The current study also demonstrated that eIF4A1 overexpression increased the NF-κB levels, whereas eIF4A1 knockdown decreased NF-κB expression in GC cells. These results indicated that eIF4A1 could enhance the secretion of VEGFA through the NF-κB signaling pathway, and thus facilitate angiogenesis and GC progression.
Although some advances have been achieved in the present study, several limitations remain. First, in the detection of eIF4A1 expression levels across clinical samples, the present study lacked normal gastric mucosal tissues as a control group, which may compromise the interpretability of comparative analyses. Second, the mechanistic relationship between eIF4A1 and immune cell infiltration/function requires more rigorous validation, particularly through multi-omics approaches and functional assays. Third, current investigations into eIF4A1-mediated regulation of the NF-κB/VEGFA signaling axis remain at a preliminary stage, with insufficient exploration of upstream regulatory nodes and downstream effector pathways. Addressing these deficiencies will constitute the core focus of our future direction.
In conclusion, the present study demonstrated that eIF4A1 was highly expressed in GC tissue compared with in pericarcinomatous tissue. High eIF4A1 expression was associated with clinicopathological factors and poor prognosis. Furthermore, it was suggested that eIF4A1 might promote tumor immune escape in GC by affecting immune cells, and that it could promote cancer angiogenesis through the VEGFA/NF-κB pathway. Therefore, eIF4A1 has the potential to act as a prognostic biomarker and a therapeutic target for patients with GC.
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from the Autonomous Research Project of Anhui Provincial Key Laboratory on Active Biomacromolecules (grant no. LAB202005), and the National Innovation and Entrepreneurship Training for the University of PRC (grant no. 202110368068).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
XZ, LJ, QT, XC and XZ performed the data curation and formal analysis. XZ, LJ and XK conducted the investigation and wrote the original draft. BC, HZ, HH and XH were responsible for statistical analysis and prepared all figures for publication. ZF and WH were responsible for study conceptualization, funding acquisition, project administration, supervision and reviewing and editing the manuscript. XZ and WH confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Nanjing Medical University [(2017) approval no. 335]. Written informed consent was obtained from all patients enrolled in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–49. 2021.PubMed/NCBI | |
|
de Martel C, Georges D, Bray F, Ferlay J and Clifford GM: Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob Health. 8:e180–e190. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Li ZY, Liu K and Zhang WH: Epidemiologic features and trends of gastric cancer in the world and China:interpretation of the GLOBOCAN 2018–2022. Chin J Bases Clin Gen Surg. 31:1236–1245. 2024.https://www.gensurg.cn/article/10.7507/1007-9424.202409074 | |
|
Kuai X, Jia L, Yang T, Huang X, Zhao W, Zhang M, Chen Y, Zhu J, Feng Z and Tang Q: Trop2 promotes multidrug resistance by regulating notch1 signaling pathway in gastric cancer cells. Med Sci Monit. 26:e9195662020. View Article : Google Scholar : PubMed/NCBI | |
|
Jungers CF, Elliff JM, Masson-Meyers DS, Phiel CJ and Origanti S: Regulation of eukaryotic translation initiation factor 6 dynamics through multisite phosphorylation by GSK3. J Biol Chem. 295:12796–12813. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Raza F, Waldron JA and Quesne JL: Translational dysregulation in cancer: eIF4A isoforms and sequence determinants of eIF4A dependence. Biochem Soc Trans. 43:1227–1233. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Lu W, Wilczynska A, Smith E and Bushell M: The diverse roles of the eIF4A family: You are the company you keep. Biochem Soc Trans. 42:166–172. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Galicia-Vázquez G, Cencic R, Robert F, Agenor AQ and Pelletier J: A cellular response linking eIF4AI activity to eIF4AII transcription. RNA. 18:1373–1384. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Williams-Hill DM, Duncan RF, Nielsen PJ and Tahara SM: Differential expression of the murine eukaryotic translation initiation factor isogenes eIF4A(I) and eIF4A(II) is dependent upon cellular growth status. Arch Biochem Biophys. 338:111–120. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Fang D, Peng J, Wang G, Zhou D and Geng X: Upregulation of eukaryotic translation initiation factor 4E associates with a poor prognosis in gallbladder cancer and promotes cell proliferation in vitro and in vivo. Int J Mol Med. 44:1325–1332. 2019.PubMed/NCBI | |
|
Horvilleur E, Sbarrato T, Hill K, Spriggs RV, Screen M, Goodrem PJ, Sawicka K, Chaplin LC, Touriol C, Packham G, et al: A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia. 28:1092–1102. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al: RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 513:65–70. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Wiegering A, Uthe FW, Jamieson T, Ruoss Y, Hüttenrauch M, Küspert M, Pfann C, Nixon C, Herold S, Walz S, et al: Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high MYC levels in colorectal cancer. Cancer Discov. 5:768–781. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J and Mori N: Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol. 81:713–722. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Wu YT, Xin L, Lu LJ, Gan L, Dai W, Shi YL, Adhikari VP, Wu KN and Kong LQ: Effect of neoadjuvant chemotherapy on the expression of hormone receptors and Ki-67 in Chinese breast cancer patients: A retrospective study of 525 patients. J Biomed Res. 32:191–197. 2017.(Epub ahead of print). PubMed/NCBI | |
|
Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al: Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 5:e10006762009. View Article : Google Scholar : PubMed/NCBI | |
|
Yoon SJ, Park J, Shin Y, Choi Y, Park SW, Kang SG, Son HY and Huh YM: Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. BMC Cancer. 20:3142020. View Article : Google Scholar : PubMed/NCBI | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI | |
|
Gao C, Guo X, Xue A, Ruan Y, Wang H and Gao X: High intratumoral expression of eIF4A1 promotes epithelial-to-mesenchymal transition and predicts unfavorable prognosis in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 52:310–319. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Huang C, Li H, Xu Y, Xu C, Sun H, Li Z, Ge Y, Wang H, Zhao T, Gao S, et al: BICC1 drives pancreatic cancer progression by inducing VEGF-independent angiogenesis. Signal Transduct Target Ther. 8:3794–3805. 2023.PubMed/NCBI | |
|
García-García C, Frieda KL, Feoktistova K, Fraser CS and Block SM: RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science. 348:1486–1488. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Fu XD: RNA helicases regulate RNA condensates. Cell Res. 30:281–282. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, Miller J, Gräf S, Provenzano E, Blows F, et al: The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 6:e16032015. View Article : Google Scholar : PubMed/NCBI | |
|
Jin C, Rajabi H, Rodrigo CM, Porco JA Jr and Kufe D: Targeting the eIF4A RNA helicase blocks translation of the MUC1-C oncoprotein. Oncogene. 32:2179–2188. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Cencic R, Robert F, Galicia-Vázquez G, Malina A, Ravindar K, Somaiah R, Pierre P, Tanaka J, Deslongchamps P and Pelletier J: Modifying chemotherapy response by targeted inhibition of eukaryotic initiation factor 4A. Blood Cancer J. 3:e1282013. View Article : Google Scholar : PubMed/NCBI | |
|
Zhao W, Zhu H, Zhang S, Yong H, Wang W, Zhou Y, Wang B, Wen J, Qiu Z, Ding G, et al: Trop2 is overexpressed in gastric cancer and predicts poor prognosis. Oncotarget. 7:6136–6145. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Cui JX, Xu XH, He T, Liu JJ, Xie TY, Tian W and Liu JY: L-kynurenine induces NK cell loss in gastric cancer microenvironment via promoting ferroptosis. J Exp Clin Cancer Res. 42:522023. View Article : Google Scholar : PubMed/NCBI | |
|
Choo J, Kua LF, Soe MY, Asuncion BR, Tan BKJ, Teo CB, Tay RYK, So J, Shabbir A, Guowei K, et al: Clinical relevance of PD-1 positive CD8 T-cells in gastric cancer. Gastric Cancer. 26:393–404. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Lundberg A, Li B and Li R: B cell-related gene signature and cancer immunotherapy response. Br J Cancer. 126:899–906. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Qu Y, Wang X, Bai S, Niu L, Zhao G, Yao Y, Li B and Li H: The effects of TNF-α/TNFR2 in regulatory T cells on the microenvironment and progression of gastric cancer. Int J Cancer. 150:1373–1391. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Su H, Jin Y, Tao C, et al: Th2 cells infiltrating high-grade serous ovarian cancer: a feature that may account for the poor prognosis. J Gynecol Oncol. 34:e482023. View Article : Google Scholar : PubMed/NCBI | |
|
Hsieh HL and Tsai MM: Tumor progression-dependent angiogenesis in gastric cancer and its potential application. World J Gastrointest Oncol. 11:686–704. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Fane ME, Ecker BL, Kaur A, Marino GE, Alicea GM, Douglass SM, Chhabra Y, Webster MR, Marshall A, Colling R, et al: sFRP2 supersedes VEGF as an age-related driver of angiogenesis in melanoma, affecting response to anti-VEGF therapy in older patients. Clin Cancer Res. 26:5709–5719. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Wei W, Cao W, Zhan Z, Yan L, Xie Y and Xiao Q: MiR-1284 suppresses gastric cancer progression by targeting EIF4A1. Onco Targets Ther. 12:3965–3976. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Ghalehbandi S, Yuzugulen J, Pranjol MZI and Pourgholami MH: The role of VEGF in cancer-induced angiogenesis and research progress of drugs targeting VEGF. Eur J Pharmacol. 949:1755862023. View Article : Google Scholar : PubMed/NCBI | |
|
Patel SA, Nilsson MB, Le X, Cascone T, Jain RK and Heymach JV: Molecular mechanisms and future implications of VEGF/VEGFR in cancer therapy. Clin Cancer Res. 29:30–39. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Jiang Y, Huang J, Huang Z, Li W, Tan R, Li T, Chen Z, Tang X, Zhao Y, Qiu J, et al: ADAMTS12 promotes oxaliplatin chemoresistance and angiogenesis in gastric cancer through VEGF upregulation. Cell Signal. 111:1108662023. View Article : Google Scholar : PubMed/NCBI | |
|
Wang R, Ma Y, Zhan S, Zhang G, Cao L, Zhang X, Shi T and Chen W: B7-H3 promotes colorectal cancer angiogenesis through activating the NF-κB pathway to induce VEGFA expression. Cell Death Dis. 11:552020. View Article : Google Scholar : PubMed/NCBI | |
|
Mantsounga CS, Lee C, Neverson J, Sharma S, Healy A, Berus JM, Parry C, Ceneri NM, López-Giráldez F, Chun HJ, et al: Macrophage IL-1β promotes arteriogenesis by autocrine STAT3- and NF-κB-mediated transcription of pro-angiogenic VEGF-A. Cell Rep. 38:1103092022. View Article : Google Scholar : PubMed/NCBI | |
|
Li F and Zhu W: LINC00460 promotes angiogenesis by enhancing NF-κB-mediated VEGFA expression in cervical cancer cells. Biochem Biophys Res Commun. 671:146–152. 2023. View Article : Google Scholar : PubMed/NCBI |