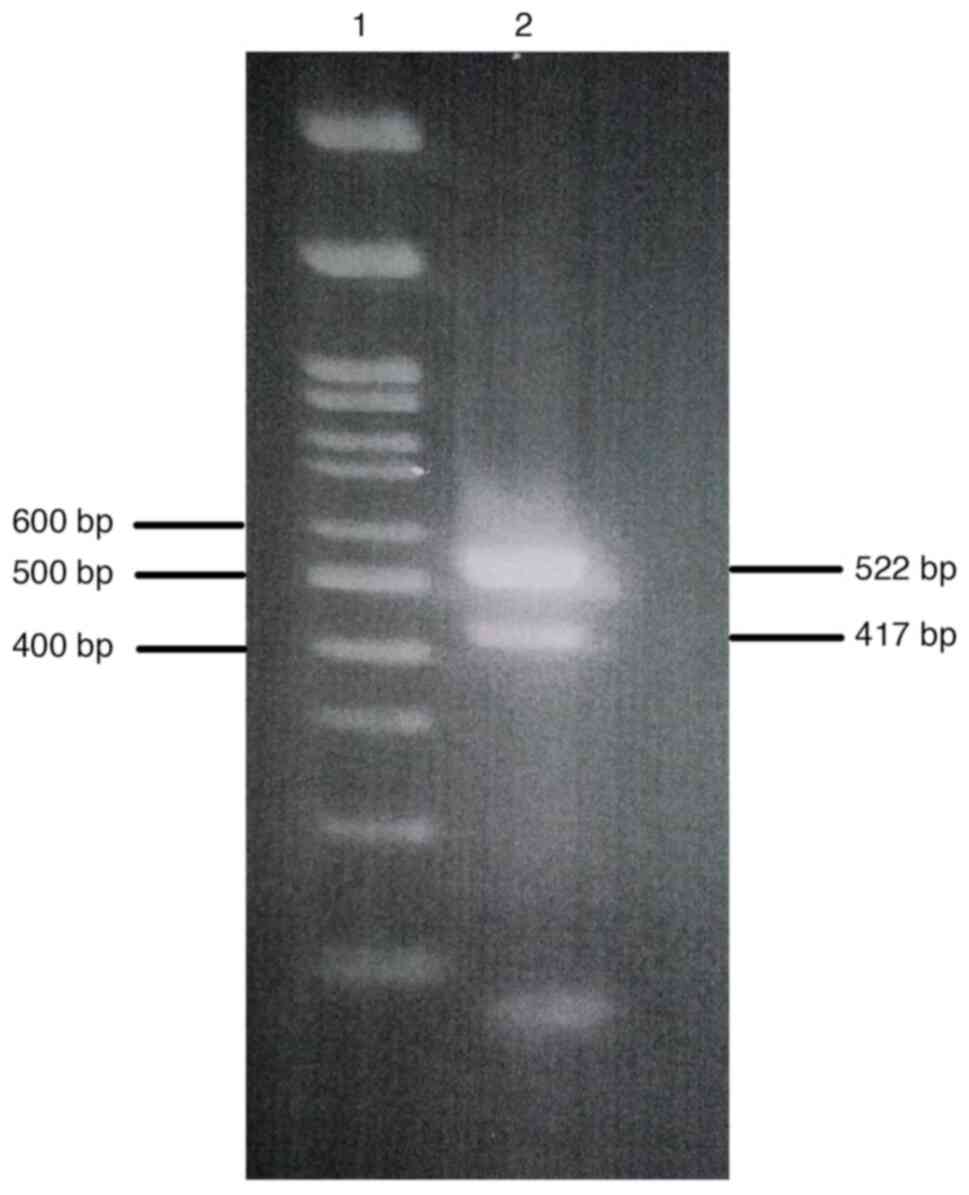
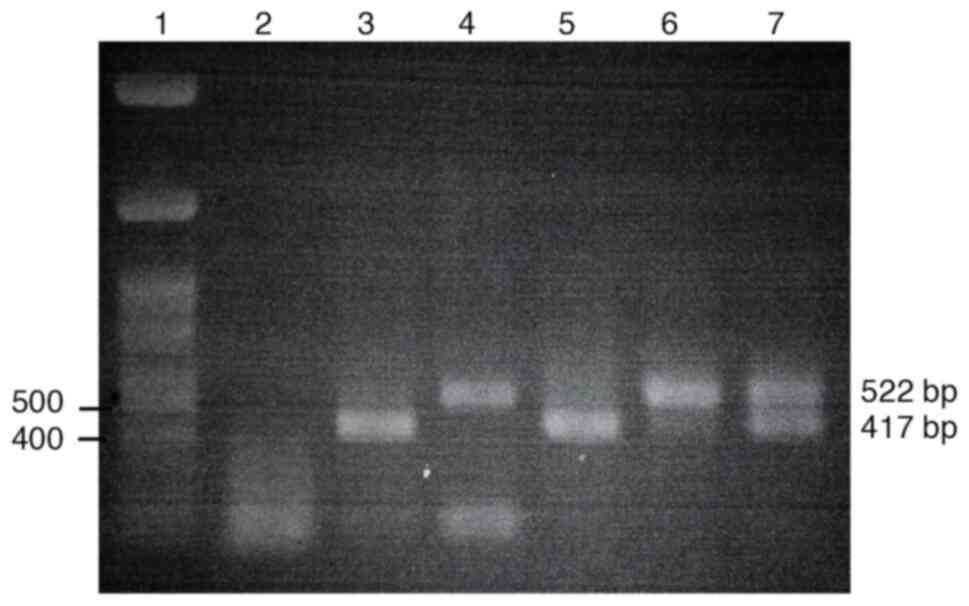
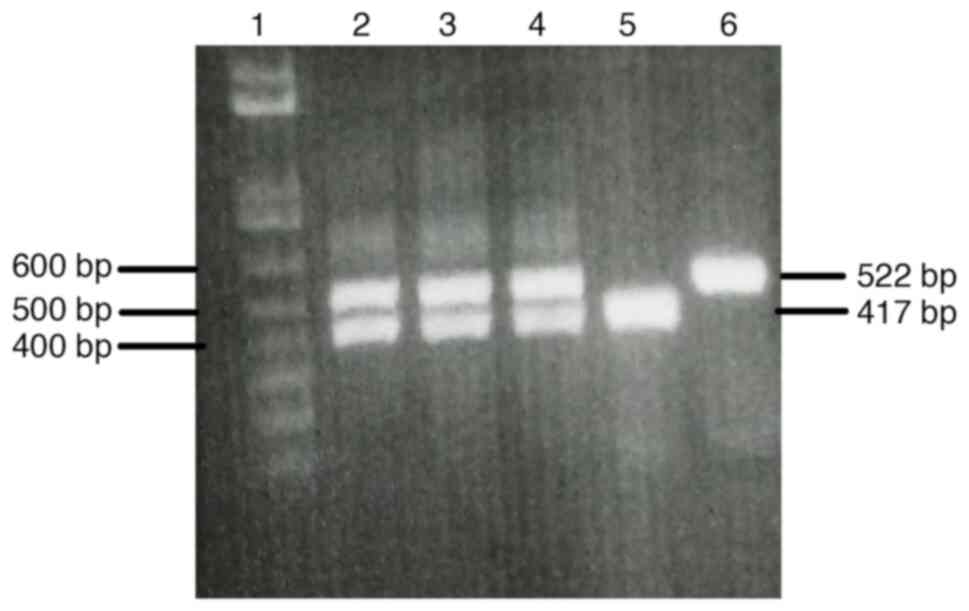
|
1
|
Polaris Observatory HCV Collaborators.
Global change in hepatitis C virus prevalence and cascade of care
between 2015 and 2020: A modelling study. Lancet Gastroenterol
Hepatol. 7:396–415. 2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Polaris Observatory HCV Collaborators.
Global prevalence and genotype distribution of hepatitis C virus
infection in 2015: A modelling study. Lancet Gastroenterol Hepatol.
2:161–176. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bochud PY, Cai T, Overbeck K, Bochud M,
Dufour JF, Müllhaupt B, Borovicka J, Heim M, Moradpour D, Cerny A,
et al: Genotype 3 is associated with accelerated fibrosis
progression in chronic hepatitis C. J Hepatol. 51:655–666.
2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nkontchou G, Ziol M, Aout M, Lhabadie M,
Baazia Y, Mahmoudi A, Roulot D, Ganne-Carrie N, Grando-Lemaire V,
Trinchet JC, et al: HCV genotype 3 is associated with a higher
hepatocellular carcinoma incidence in patients with ongoing viral C
cirrhosis. J Viral Hepat. 18:e516–e522. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sorbo MC, Cento V, Di Maio VC, Howe AYM,
Garcia F, Perno CF and Ceccherini-Silberstein F: Hepatitis C virus
drug resistance associated substitutions and their clinical
relevance: Update 2018. Drug Resist Updat. 37:17–39.
2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
da Silva DL, Nunes HM and Freitas PEB:
Natural prevalence of NS3 gene resistance-associated substitutions
(RASs) in patients with chronic hepatitis C from the state of
Pará/Brazil. Virus Res. 292(198251)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Li Z, Chen ZW, Li H, Ren H and Hu P:
Prevalence of hepatitis C virus-resistant association substitutions
to direct-acting antiviral agents in treatment-naïve hepatitis C
genotype 1b-infected patients in western China. Infect Drug Resist.
10:377–392. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen ZW, Li H, Ren H and Hu P: Global
prevalence of preexisting HCV variants resistant to direct-acting
antiviral agents (DAAs): mining the GenBank HCV genome data. Sci
Rep. 6(20310)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zeuzem S, Mizokami M, Pianko S, Mangia A,
Han KH, Martin R, Svarovskaia E, Dvory-Sobol H, Doehle B, Hedskog
C, et al: NS5A resistance-associated substitutions in patients with
genotype 1 hepatitis C virus: prevalence and effect on treatment
outcome. J Hepatol. 66:910–918. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mushtaq S, Hashmi AH, Khan A, Asad Raza
Kazmi SM and Manzoor S: Emergence and persistence of
resistance-associated substitutions in HCV GT3 patients failing
direct-acting antivirals. Front Pharmacol.
13(894460)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Smith D, Magri A, Bonsall D, Ip CLC,
Trebes A, Brown A, Piazza P, Bowden R, Nguyen D, Ansari MA, et al:
Resistance analysis of genotype 3 hepatitis C virus indicates
subtypes inherently resistant to nonstructural protein 5A
inhibitors. Hepatology. 69:1861–1872. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bertoli A, Sorbo MC, Aragri M, Lenci I,
Teti E, Polilli E, Di Maio VC, Gianserra L, Biliotti E, Masetti C,
et al: Prevalence of single and multiple natural NS3, NS5A and NS5B
resistance-associated substitutions in hepatitis C virus genotypes
1-4 in Italy. Sci Rep. 8(8988)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kiattanaphon A, Vipsoongnern Y, Kunthalert
D and Sistayanarain A: Partial nonstructural 3 region analysis of
hepatitis C virus genotype 3a. Mol Biol Rep. 49:9437–9443.
2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mundim AEFM, de Castro FOF, Albuquerque
MBB, Vilanova-Costa CAST, Pfrimer IAH and Silva AMTC: Major
mutations in the NS3 gene region of hepatitis C virus related to
the resistance to direct acting antiviral drugs: A systematic
review. Virusdisease. 31:220–228. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shepherd SJ, Abdelrahman T, MacLean AR,
Thomson EC, Aitken C and Gunson RN: Prevalence of HCV NS3
pre-treatment resistance associated amino acid variants within a
Scottish cohort. J Clin Virol. 65:50–53. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kumthip K and Maneekarn N: The role of HCV
proteins on treatment outcomes. Virol J. 12(217)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ceccherini-Silberstein F, Cento V, Di Maio
VC, Perno CF and Craxì A: Viral resistance in HCV infection. Curr
Opin Virol. 32:115–127. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chevaliez S, Rodriguez C and Pawlotsky JM:
New virologic tools for management of chronic hepatitis B and C.
Gastroenterology. 142:1303–1313.e1. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen Q, Belmonte I, Buti M, Nieto L,
Garcia-Cehic D, Gregori J, Perales C, Ordeig L, Llorens M, Soria
ME, et al: New real-time-PCR method to identify single point
mutations in hepatitis C virus. World J Gastroenterol.
22:9604–9612. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Nguyen T, Akhavan S, Caby F, Bonyhay L,
Larrouy L, Gervais A, Lebray P, Poynard T, Calmus Y, Simon A, et
al: Net emergence of substitutions at position 28 in NS5A of
hepatitis C virus genotype 4 in patients failing direct-acting
antivirals detected by next-generation sequencing. Int J Antimicrob
Agents. 53:80–83. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wilber E, Rebolledo PA, Kasinathan V,
Merritt S, Titanji BK, Aldred B, Kandiah S, Ray SM, Sheth AN,
Colasanti JA and Wang YF: Utility of a viral vesicular panel
multiplex polymerase chain reaction assay for the diagnosis of
monkeypox, herpes simplex, and varicella zoster viruses. Open Forum
Infect Dis. 10(ofad140)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yan S, Yang F, Yao H, Dong D, Wu D, Wu N,
Ye C and Wu H: A multiplex real-time RT-PCR assay for the detection
of H1, H2 and H3 subtype avian influenza viruses. Virus Genes.
59:333–337. 2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Strommenger B, Kettlitz C, Werner G and
Witte W: Multiplex PCR assay for simultaneous detection of nine
clinically relevant antibiotic resistance genes in Staphylococcus
aureus. J Clin Microbiol. 41:4089–4094. 2003.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Coelho C, Vinatea CE, Heinert AP, Simões
CM and Barardi CR: Comparison between specific and multiplex
reverse transcription-polymerase chain reaction for detection of
hepatitis A virus, poliovirus and rotavirus in experimentally
seeded oysters. Mem Inst Oswaldo Cruz. 98:465–468. 2003.PubMed/NCBI View Article : Google Scholar
|