Amplification of NS3 and NS5A in hepatitis C virus by the multiplex nested polymerase chain reaction
- Authors:
- Published online on: April 7, 2025 https://doi.org/10.3892/wasj.2025.342
- Article Number: 54
-
Copyright : © Sistayanarain et al. This is an open access article distributed under the terms of Creative Commons Attribution License [CC BY 4.0].
Abstract
Introduction
The hepatitis C virus (HCV), a member of the Flaviviridae family, is a major cause of hepatitis, hepatic cirrhosis and hepatocellular carcinoma. The HCV genome has ~9,600 nucleotides which encode the protein core (C), envelopes (E) 1 and 2, p7, non-structural (NS) 2, NS3, NS4A, NS4B, NS5A and NS5B. Of note, ~0.7% of the worldwide population is infected with HCV (1). HCV has been grouped into eight main genotypes (types 1-8) and several subtypes, and these are unevenly distributed through various geographical regions. HCV genotype 1 is prevalent worldwide, whereas genotype 3 is mostly distributed in South Asia and Southeast Asia. Infection with HCV genotype 3 is mostly detected in lower middle-income countries (2). HCV genotype 3 has been shown to be a risk factor for the accelerated fibrosis progression of chronic hepatitis C and it is also associated with a higher incidence of hepatocellular carcinoma (3,4). During anti-HCV drug therapy, genotype 3-infected patients, particularly those with cirrhosis, are more difficult to treat than patients infected with other HCV genotypes. Direct-acting antiviral (DAA) agents have been developed for treatment regimens against HCV infection, and these function by blocking the life cycle of the virus. DAAs can be divided into three classes: Non-structural (NS3) protease inhibitors, NS5B polymerase inhibitors and NS5A inhibitors. Currently, DAA combination therapies are recommended and clinically used regardless of HCV genotypes and subtypes. Resistance-associated substitutions (RASs) result from mutations in the proteins targeted by DAAs (the NS3, NS5A and NS5B regions) and RASs decrease antiviral efficacy of DAA treatments (5). Several prevalence studies have demonstrated that mutations in the NS3 and NS5A regions are associated with a significant decrease in the sensitivity of HCV to the effects of DAAs (6-12). Specifically, the presence of the following major amino acid substitutions in the NS3 gene region is associated with resistance to DAAs: V36L, T54S, V55A, Q80K, S122T, R155K, A156S, 158, D168Q, V170I, N174I and M175L. The Q80K polymorphism, a NS3 resistance-associated substitution, is the most frequently detected (6,7,13-15). Amino acid mutations at positions 30, 31,58, 62 and 93 of HCV genotype 3 confer resistance to NS5A inhibitors (11,12,16,17). The identification of such NS3 and NS5A mutations is considered of critical importance for optimizing effective therapies. The method of detecting HCV mutations in patients who have failed DAA treatment and in treatment-naïve patients is traditional Sanger sequencing, a technique with a simple interpretation that is most commonly used (18). The HCV target regions are normally prepared separately using polymerase chain reaction (PCR). Later, the amplified product is used for the Sanger sequencing process, followed by identification of the amino acid mutations. Each specific HCV fragment is generally prepared using reverse transcription and then nested PCR. However, this method is time-consuming, costly and it is vulnerable to carryover contamination, which causes trouble. To improve on this, the aim of the present study was to simultaneously amplify both NS3 and NS5A of HCV by using multiplex nested PCR. Optimal primers and conditions for multiplex nested PCR were determined beforehand.
Materials and methods
Designing candidate primers for optimizing the amplification of NS3 and NS5A
Specific candidate primers for optimization of the target amplification were designed based on conserved motifs within the NS3 and NS5A regions of HCV using the Primer3 program (https://primer3.org/). The two programs were then applied to determine the specificity for NS3 and NS5A of HCV genotype 3a. The first was the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI), part of the US National Library of Medicine (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The other was the Multiple Sequence Alignment program from European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Samples
HCV-infected serum samples were obtained from the Phitsanulok Blood Center in Phitsanulok City, Thailand. The serum samples were collected from December, 2020 to August, 2021. The study protocols and procedures were approved by the Naresuan University Institutional Review Board (IRB No. P1-0134/2565). As the samples were obtained from a blood bank, it was deemed by the committee that written informed consent was not required from the patients as all samples were de-identified samples.
HCV RNA extraction and reverse transcription
The extraction of the HCV RNA from the serum samples was performed using the PureLink™ Viral RNA/DNA Mini kit (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into complementary DNA (cDNA) using random hexamers (5'-NNNNNN-3') and RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas; Thermo Fisher Scientific, Inc.).
Optimization of multiplex nested PCR
Multiplex nested PCR was optimized for the simultaneous amplification of both NS3 and NS5A. The first and second rounds of PCR were performed in 25 µl of the reaction mixture, which was comprised of DNA, 1X PCR buffer, dNTP (200 µM), Taq DNA polymerase (1.25 units) (Bio-Helix Co., Ltd.) and both NS3 primer (0.5 µM) and NS5A primer (0.5 µM). Details of the primer sequences are presented in Table I. For the negative control, distilled water was used. The cycling conditions for the first round were as follows: A pre-incubation step at 94˚C for 7 min, followed by 40 cycles of denaturation at 94˚C for 10 sec, annealing at 45˚C for 30 sec, extension at 68˚C for 40 sec, and then a final extension step at 68˚C for 7 min. The cycling conditions for the second round of PCR were as follows: A pre-incubation step at 94˚C for 7 min, followed by 40 cycles of denaturation at 94˚C for 30 sec, annealing at 46˚C for 30 sec, extension at 72˚C for 40 sec, and then a final extension step at 72˚C for 7 min. The amplified products of the NS3 and NS5A regions were identified using 1.0% agarose gel electrophoresis. The results of the amplified product identification were used to determine the optimal primers and conditions for multiplex nested PCR.
Evaluation of multiplex nested PCR
Using the most effective primers and conditions thus determined, the optimized multiplex RT-PCR assay was then carried out on all 20 serum samples.
Results
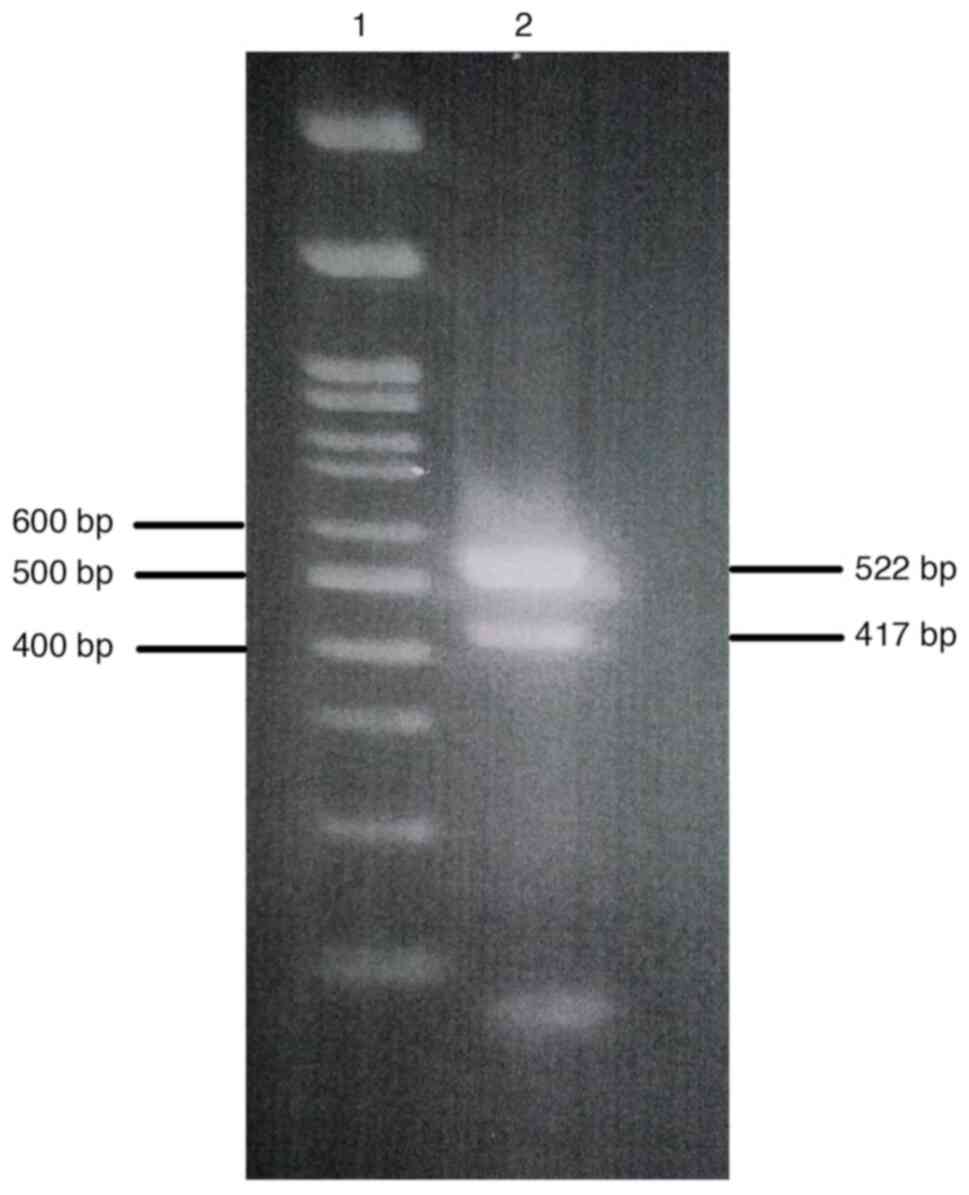
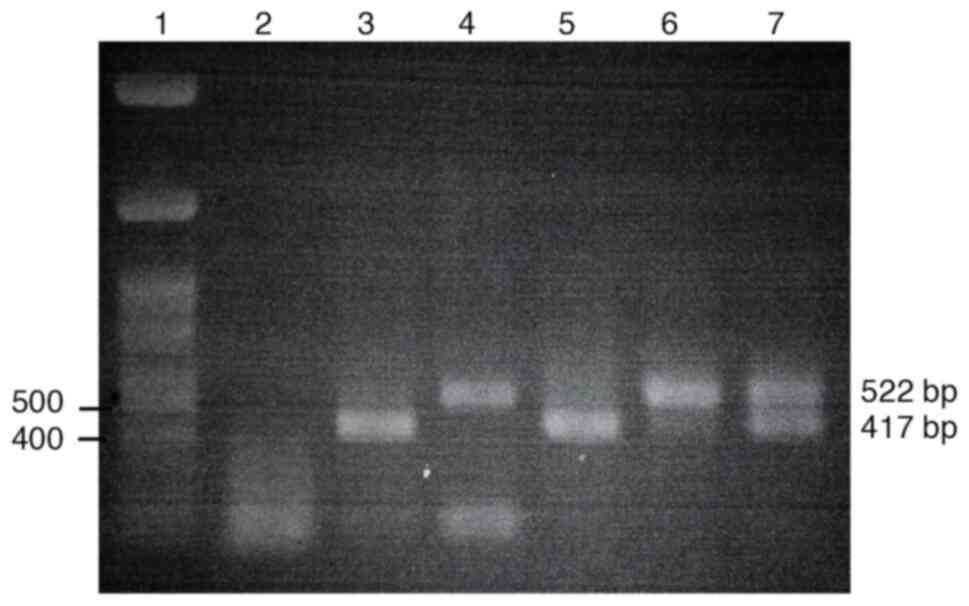
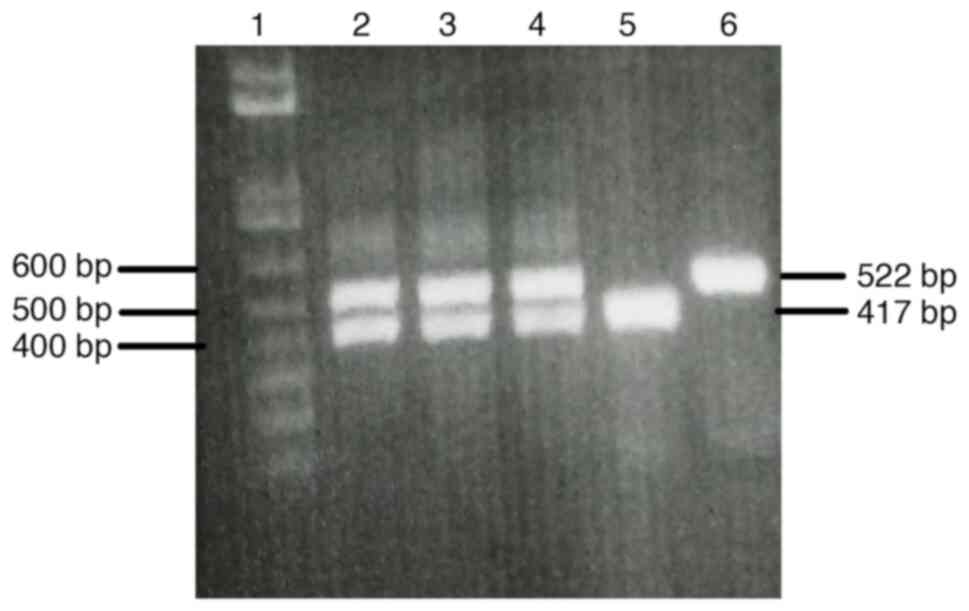
The initial experiment was run to optimize the primers and assay conditions for use in multiplex nested PCR to amplify product fragments of both NS3 and NS5A (data not shown). The multiplex nested PCR results were considered positive if specific amplicons could be obtained for both the NS3 and the NS5A regions. The amplification results represented the PCR product fragments of the NS3 and the NS5A as 417 and 522 bp. A cDNA was also used as a positive template to further optimize the multiplex nested PCR conditions, and the results are shown in Fig. 1. Once the optimal primers and assay conditions were established, the feasibility of applying these optimizations to clinical samples was verified by evaluating fresh serum samples with optimized multiplex nested PCR. The amplification results of NS3 and NS5A from the serum samples using the optimized multiplex nested PCR are shown in Fig. 2. The amplification results of both the NS3 and NS5A regions are summarized in Table II. Within the total of 20 HCV infected serum samples, a total of 14 samples (70%) were positive for both NS3 and NS5A after employing multiplex nested PCR. Amplification results of 3 samples (15%) and 2 samples (10%) were positive for only the NS3 or NS5A amplified products, respectively. Only one sample (5%) had a negative result for both amplified targets. The cDNA template was diluted to determine the limit of detection. Both specific amplified fragments of the NS3 and NS5A were detectable at the dilution of 1/2-1/8 (Fig. 3).
Discussion
DAAs have been used in the treatment regimens for hepatitis caused by HCV infection, and it has been demonstrated that amino acid mutations in the NS3 and NS5A regions can cause resistance to DAA treatment. As the guidelines for the selection of effective DAA treatment include the study of amino acid mutation prevalence, there is a need to verify the presence or absence of the amino acid mutations in NS3 and NS5A proteins. Numerous methods have been developed for detecting mutants. The standard method for detecting mutations in the amino acids of HCV protein includes synthesizing cDNA, performing a nested PCR, purifying an amplified product fragment, and sequencing the nucleotides and amino acids. A low-cost and sensitive real time-PCR technique has been successfully developed to identify resistance-associated mutations in HCV infected samples, but it only works for point mutations (19). Next-generation sequencing (NGS) is a sensitive technique for detection of minority RASs (20). However, NGS is a costly technique that is not widely used. Direct Sanger sequencing is an easy-to-use and inexpensive method that can detect a number of mutated amino acid positions. The specific PCR fragment gene is first prepared for nucleotide and amino acid sequencing. To detect mutations of multiple genes, the genes to be amplified are normally processed in separate PCR tubes with different conditions. However, each time that PCR is performed, there is a danger of serious contamination leading to erroneous results. Thus, carrying out PCR in two tubes doubles the chance of contamination. By contrast, the present study investigated preparing the NS3 and NS5B fragments by using the multiplex PCR method, which has the potential to amplify multiple gene targets in a single PCR run. Multiplex PCR assays have previously been applied for simultaneous detection of several viral infections, including detection of antibiotic resistance genes and pathogen subtypes (21-24). The aim in the present study was to optimize the simultaneous amplification of NS3 and NS5A of HCV for a rapid, specific and inexpensive multiplex nested PCR technique. The set of candidate primers to amplify the NS3 and NS5A were designed based on the nucleotide sequence of HCV genotype 3a. In the preliminary experiments, positive PCR fragments were sought in order to verify the optimal conditions for the primary multiplex PCR. Amplification failure occurred when using some positive PCR fragments templates (data not shown). Later, a cDNA positive template was utilized to further optimize the final multiplex nested PCR conditions. The 5 µl of cDNA template is the optimum volume for simultaneous amplification of NS3 and NS5A regions. Both amplified NS3 and NS5A were detected in 14 out of 20 serum samples (70%) through the multiplex nested PCR system, while only one sample had a negative detection of both genes. One possible explanation for the results that did not detect both NS3 and NS5A is that the effectiveness of the set of primers used could have been hindered by NS3 and/or NS5A variation. The analytical limit of simultaneous amplification of NS3 and NS5A was the 1/8 dilution of the initial 5 µl of cDNA template. When the cDNA template concentration was low, either the NS3 amplicon or the NS5A amplicon was observed (Fig. 3). The exact concentration of starting cDNA, which was not measured, is one of the limitations of the present study. This is a preliminary optimized technique to amplify both NS3 and NS5A of HCV genotype 3a in single-tube multiplex nested PCR. It is possible that this method could be applied to achieve simultaneous amplification of other HCV genotypes, as well. In addition, this technique could be adapted for the simultaneous amplification of multiple targets, e.g. NS3, NS5A and NS5B. In the present study, the multiplex nested PCR technique for the simultaneous amplification of NS3 and NS5A of HCV was successfully designed and its working conditions optimized. In conclusion, multiplex nested PCR is a rapid, effective, sensitive and inexpensive method for use in the simultaneous amplification of NS3 and NS5A.
Acknowledgements
The authors would like to thank the Phitsanulok Blood Center in Phitsanulok City, Thailand for providing the serum samples. The authors would also like to extend their gratitude to Mr. Paul Freund of Naresuan University Writing Clinic (DIALD) for providing editing assistance.
Funding
Funding: The present study was funded by the National Science Research and Innovation Fund (NSRF), Year 2023 (Fundamental Fund/Grant R2566B056).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
AS was involved in the conception and design of the study. Both authors (AS and DK) were involved in the study methodology, data collection, data analysis and writing the manuscript. Both authors (AS and DK) have read and approved the manuscript for the publication. Both authors (AS and DK) confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The study protocols and procedures were approved by the Naresuan University Institutional Review Board (IRB No. P1-0134/2565). As the samples were obtained from a blood bank, it was deemed by the committee that written informed consent was not required from the patients as all samples were de-identified samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Polaris Observatory HCV Collaborators. Global change in hepatitis C virus prevalence and cascade of care between 2015 and 2020: A modelling study. Lancet Gastroenterol Hepatol. 7:396–415. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Polaris Observatory HCV Collaborators. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: A modelling study. Lancet Gastroenterol Hepatol. 2:161–176. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Bochud PY, Cai T, Overbeck K, Bochud M, Dufour JF, Müllhaupt B, Borovicka J, Heim M, Moradpour D, Cerny A, et al: Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J Hepatol. 51:655–666. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Nkontchou G, Ziol M, Aout M, Lhabadie M, Baazia Y, Mahmoudi A, Roulot D, Ganne-Carrie N, Grando-Lemaire V, Trinchet JC, et al: HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepat. 18:e516–e522. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Sorbo MC, Cento V, Di Maio VC, Howe AYM, Garcia F, Perno CF and Ceccherini-Silberstein F: Hepatitis C virus drug resistance associated substitutions and their clinical relevance: Update 2018. Drug Resist Updat. 37:17–39. 2018.PubMed/NCBI View Article : Google Scholar | |
|
da Silva DL, Nunes HM and Freitas PEB: Natural prevalence of NS3 gene resistance-associated substitutions (RASs) in patients with chronic hepatitis C from the state of Pará/Brazil. Virus Res. 292(198251)2021.PubMed/NCBI View Article : Google Scholar | |
|
Li Z, Chen ZW, Li H, Ren H and Hu P: Prevalence of hepatitis C virus-resistant association substitutions to direct-acting antiviral agents in treatment-naïve hepatitis C genotype 1b-infected patients in western China. Infect Drug Resist. 10:377–392. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Chen ZW, Li H, Ren H and Hu P: Global prevalence of preexisting HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep. 6(20310)2016.PubMed/NCBI View Article : Google Scholar | |
|
Zeuzem S, Mizokami M, Pianko S, Mangia A, Han KH, Martin R, Svarovskaia E, Dvory-Sobol H, Doehle B, Hedskog C, et al: NS5A resistance-associated substitutions in patients with genotype 1 hepatitis C virus: prevalence and effect on treatment outcome. J Hepatol. 66:910–918. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Mushtaq S, Hashmi AH, Khan A, Asad Raza Kazmi SM and Manzoor S: Emergence and persistence of resistance-associated substitutions in HCV GT3 patients failing direct-acting antivirals. Front Pharmacol. 13(894460)2022.PubMed/NCBI View Article : Google Scholar | |
|
Smith D, Magri A, Bonsall D, Ip CLC, Trebes A, Brown A, Piazza P, Bowden R, Nguyen D, Ansari MA, et al: Resistance analysis of genotype 3 hepatitis C virus indicates subtypes inherently resistant to nonstructural protein 5A inhibitors. Hepatology. 69:1861–1872. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Bertoli A, Sorbo MC, Aragri M, Lenci I, Teti E, Polilli E, Di Maio VC, Gianserra L, Biliotti E, Masetti C, et al: Prevalence of single and multiple natural NS3, NS5A and NS5B resistance-associated substitutions in hepatitis C virus genotypes 1-4 in Italy. Sci Rep. 8(8988)2018.PubMed/NCBI View Article : Google Scholar | |
|
Kiattanaphon A, Vipsoongnern Y, Kunthalert D and Sistayanarain A: Partial nonstructural 3 region analysis of hepatitis C virus genotype 3a. Mol Biol Rep. 49:9437–9443. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Mundim AEFM, de Castro FOF, Albuquerque MBB, Vilanova-Costa CAST, Pfrimer IAH and Silva AMTC: Major mutations in the NS3 gene region of hepatitis C virus related to the resistance to direct acting antiviral drugs: A systematic review. Virusdisease. 31:220–228. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Shepherd SJ, Abdelrahman T, MacLean AR, Thomson EC, Aitken C and Gunson RN: Prevalence of HCV NS3 pre-treatment resistance associated amino acid variants within a Scottish cohort. J Clin Virol. 65:50–53. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Kumthip K and Maneekarn N: The role of HCV proteins on treatment outcomes. Virol J. 12(217)2015.PubMed/NCBI View Article : Google Scholar | |
|
Ceccherini-Silberstein F, Cento V, Di Maio VC, Perno CF and Craxì A: Viral resistance in HCV infection. Curr Opin Virol. 32:115–127. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Chevaliez S, Rodriguez C and Pawlotsky JM: New virologic tools for management of chronic hepatitis B and C. Gastroenterology. 142:1303–1313.e1. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Chen Q, Belmonte I, Buti M, Nieto L, Garcia-Cehic D, Gregori J, Perales C, Ordeig L, Llorens M, Soria ME, et al: New real-time-PCR method to identify single point mutations in hepatitis C virus. World J Gastroenterol. 22:9604–9612. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Nguyen T, Akhavan S, Caby F, Bonyhay L, Larrouy L, Gervais A, Lebray P, Poynard T, Calmus Y, Simon A, et al: Net emergence of substitutions at position 28 in NS5A of hepatitis C virus genotype 4 in patients failing direct-acting antivirals detected by next-generation sequencing. Int J Antimicrob Agents. 53:80–83. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Wilber E, Rebolledo PA, Kasinathan V, Merritt S, Titanji BK, Aldred B, Kandiah S, Ray SM, Sheth AN, Colasanti JA and Wang YF: Utility of a viral vesicular panel multiplex polymerase chain reaction assay for the diagnosis of monkeypox, herpes simplex, and varicella zoster viruses. Open Forum Infect Dis. 10(ofad140)2023.PubMed/NCBI View Article : Google Scholar | |
|
Yan S, Yang F, Yao H, Dong D, Wu D, Wu N, Ye C and Wu H: A multiplex real-time RT-PCR assay for the detection of H1, H2 and H3 subtype avian influenza viruses. Virus Genes. 59:333–337. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Strommenger B, Kettlitz C, Werner G and Witte W: Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 41:4089–4094. 2003.PubMed/NCBI View Article : Google Scholar | |
|
Coelho C, Vinatea CE, Heinert AP, Simões CM and Barardi CR: Comparison between specific and multiplex reverse transcription-polymerase chain reaction for detection of hepatitis A virus, poliovirus and rotavirus in experimentally seeded oysters. Mem Inst Oswaldo Cruz. 98:465–468. 2003.PubMed/NCBI View Article : Google Scholar |